Original Data
| Rev Diabet Stud,
2016,
13(1):66-78 |
DOI 10.1900/RDS.2016.13.66 |
Chronic Exposure to Proline Causes Aminoacidotoxicity and Impaired Beta-Cell Function: Studies In Vitro
Zhenping Liu1,2, Per B. Jeppesen1, Søren Gregersen1, Lotte Bach Larsen3, Kjeld Hermansen1
1Department of Medicine and Endocrinology, Aarhus University Hospital, Tage-Hansens Gade 2, DK-8000 Aarhus C, Denmark
2Department of Endocrinology, PLA 309 Hospital, 17 Heishanhu Road, Haidian District, 100091, Beijing, P. R. China
3Department of Food Science, Faculty of Agricultural Sciences, Aarhus University, DK-8230 Tjele, Denmark
Address correspondence to: Zhenping Liu, e-mail: zhengping.liu@ki.au.dk
Manuscript submitted August 31, 2015; resubmitted October 7, 2015; accepted December 12, 2015.
Keywords: type 2 diabetes, amino acid, pancreatic β-cell, cytochrome c oxidase, cholesterol biosynthesis, INS-1E, insulin
Abstract
BACKGROUND: Pancreatic islet-cell dysfunction is a hallmark in the development of diabetes, but the reasons for the primary β-cell defect are still elusive. Elevated circulating proline levels have been found in subjects with insulin resistance, obesity, and type 2 diabetes. Therefore, we assessed β-cell function, gene expressions, and cell death after long-term exposure of pancreatic β-cells to excess proline in vitro. METHODS: Isolated mouse islets and INS-1E cells were incubated with and without excess proline. After 72 h, we examined: (1) β-cell function, including basal insulin secretion (BIS) and glucose-stimulated insulin secretion (GSIS), (2) transcription factors related to insulin gene expression and enzymes involved in the tricarboxylic acid cycle and cholesterol biogenesis, (3) cellular triglycerides (TG) and cholesterol content, (4) the death of INS-1E cells and 3H thymidine incorporation, and (5) protein expression of INS-1E cells in response to proline by proteomics. RESULTS: We found that high doses of proline increased BIS and decreased GSIS in both isolated mouse islets and INS-1E cells. MafA, insulin 1, and the cytochrome c oxidase subunit VIa polypeptide 2 mRNA expressions were all downregulated, indicating that proline impaired insulin gene transcription and mitochondrial oxidative phosphorylation. In contrast, mevalonate decarboxylase gene expression was upregulated, and simultaneously, cholesterol content in INS-1E cells was enhanced. Protein profiling of INS-1E cells revealed that cytosolic non-specific dipeptidase and α enolase were differentially expressed. CONCLUSIONS: Our results indicate that proline-induced insulin transcription and mitochondrial oxidative phosphorylation impairment may contribute to the β-cell dysfunction observed in type 2 diabetes. Caution should be applied in interpreting the pathophysiological role of proline since very high proline concentrations were used in the experiments.
Abbreviations: AA amino acids; ABCA1 ATP-binding cassette transporter subfamily A member 1; ABCG1 ATP-binding cassette transporter G subfamily 1; ACC1 acetyl-CoA carboxylase α; AGE advanced glycation end-products; AMPK 5' adenosine monophosphate-activated protein kinase; BIS basal insulin secretion; Bmf Bcl2 modifying factor; BSA bovine serum albumin; Casp3 caspase-3; CH cholesterol; CHAPS 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate; Cox cytochrome c oxidase; Cox6a2 cytochrome c oxidase, subunit VIa, polypeptide 2; Dapk3 death-associated protein kinase 3; DTE dithioerythritol; Fasn fatty acid synthase; FBS fetal bovine serum; GPO-PAP glycerol-3-phosphate oxidase phenol aminophenazone; GSIS glucose-stimulated insulin secretion; INS-1E rat insulinoma 1E cell line; Ins1 insulin 1; Ins2 insulin 2; IPG immobilized pH gradient; IR insulin resistance; MafA v-maf musculoaponeurotic fibrosarcoma oncogene homolog A; M-KRB modified Krebs-Ringer buffer; Mvd mevalonate (diphospho) decarboxylase; OPA-1 optic atrophy 1; OxPhos oxidative phosphorylation; PDH pyruvate dehydrogenase; Pdk2 pyruvate dehydrogenase kinase 2; Pdx1 pancreatic and duodenal homeobox 1; PI 3-kinase phosphoinositid 3 kinase; TC tissue culture; TG triglycerides; T2D type 2 diabetes
1. Introduction
Type 2 diabetes (T2D) involves both defective islet cell function [1, 2] and insulin resistance (IR). Once diabetes is established, chronic hyperglycemia and hyperlipidemia can exert deleterious effects on β-cell function, referred to as glucotoxicity and lipotoxicity [3, 4], including increased basal insulin secretion (BIS) and impaired glucose-stimulated insulin secretion (GSIS). In the insulin-resistant state, circulating levels of amino acids (AA) are elevated, in particular proline, leucine, isoleucine, valine, and glutamine [5]. Previously, we demonstrated that chronic exposure to elevated levels of the branched amino acid leucine causes detrimental effects on both β-cell function and insulin sensitivity, i.e. aminoacidotoxicity may play a critical role in the pathogenesis of T2D [6].
Obese subjects are more insulin-resistant than lean subjects, and have higher circulating proline levels [5]. In agreement with this finding, higher levels of proline were found in Chinese individuals with increased IR [7]. Elevated AA concentrations have also been detected in skeletal muscles of diet-induced obese animals [8]. AAs exert negative effects on insulin action in muscles, and this is associated with inhibitory phosphorylation of insulin receptor substrate 1 on serine and/or threonine residues and impaired activation of PI 3-kinase [8]. The negative modulatory effects of AAs on insulin action have also been demonstrated in cultured hepatocytes [9] and adipocytes [10], which are also involved in dysregulated insulin receptor substrate-1-mediated signaling.
Previously, we have demonstrated in acute studies that proline stimulates insulin secretion in both clonal β-cells and isolated mouse islets [11, 12]. However, to the best of our knowledge, there exists no information on the long-term effect of proline on pancreatic β-cells.
Proline is an abundant AA occurring in milk proteins, meat, poultry by-products, and salmon proteins [13]. We hypothesize that chronic exposure to elevated proline levels results in changes in β-cell function characteristic for the diabetic condition, i.e. increased BIS, impaired GSIS, and altered β-cell gene expression. We assessed the impact of high concentrations of proline after 72 h culture in a clonal β-cell line, INS-1E, on the parameters of β-cell function, including insulin secretion, cell proliferation, cell death, gene expression, and protein change.
2. Materials and methods
2.1 Reagents and buffers/solutions
All chemicals were obtained from Sigma-Aldrich (Brøndy, Denmark) if not stated otherwise. Modified Krebs-Ringer buffer (M-KRB) contained the following components:
- NaCl 125 mM
- KCl 5.9 mM
- MgCl2 1.2 mM
- CaCl2 1.28 mM
- NaHCO3 5.0 mM
- Hepes: 25 mM
At pH 7.4, SYTO 24 solution contained 5 mM SYTO 24 green fluorescent nucleic acid stain (molecular probes, Invitrogen, Oregon, USA) dissolved in dimethyl sulfoxside ≥ 99.9% to a final concentration of 0.01 mM. Glycine bovine serum albumin (BSA) buffer contained glycine 100 mM and 0.25% BSA, with pH 8.8 (Roche Molecular Biochemicals, Mannheim, Germany). 2D-lysis buffer contained 6 M urea, 2 M thiourea, 1.5% (w/v) pharmalyte, 0.8% (w/v) 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS), and 1% (w/v) dithioerythritol (DTE) in water. The rehydration buffer consisted of the same substances, in the same concentrations as the lysis buffer, but with extra pharmalyte (5 μl/ml).
2.2 Experimental animals
Adult female NMRI mice (Bomholtgaard Breeding and Research Centre, Ry, Denmark) weighing 20-25 g were used. The animals were kept on a standard pellet diet, tap water ad libitum, and a 12 h/12 h light/darkness cycle. This study was carried out in accordance with the guidelines of the Danish Council for Animal Experiments.
Islet isolation. Islets were isolated by the collagenase digestion technique [14]. In brief, the animals were anesthetized intraperitoneally with pentobarbital sodium (50 mg/kg), and a midline laparotomy was performed. The pancreas was retrogradely filled with 3 ml ice-cold Hanks' balanced salt solution (HBSS), supplemented with 0.3 mg/ml collagenase P (Boehringer Mannheim, Mannheim, Germany). The pancreas was removed, incubated for 19 min at 37°C in a water bath, and subsequently rinsed with ice-cold HBSS. Subsequently, the islets were handpicked under a stereomicroscope. The islets were incubated overnight at 37°C and 95% normal atmosphere (5% CO2) in 10 ml of RPMI 1640 containing 11.1 mM glucose, supplemented with 10% fetal bovine serum (FBS), 2.06 mM glutamine, 100 IU/ml penicillin G, and 100 µg/ml streptomycin (all GIBCO-BRL, Paisley, UK).
Insulin secretion from isolated islets. After overnight incubation, mouse islets were incubated in RPMI 1640 with 11.1 mM glucose each in the presence of 1, 5, and 10 mM proline. Normal RPMI 1640 containing 0.17 mM proline was used as control. After 72 h proline treatment, islets were washed once with M-KRB, supplemented with 3.3 mM glucose and 0.1% human serum albumin, and preincubated for 30 min in M-KRB at 37°C. Subsequently, a single islet was handpicked and incubated in 100 µl M-KRB and 3.3 or 16.7 mM glucose. After 60 min of incubation in normal atmosphere at 37°C, the medium (50 µl) was collected and frozen in preparation for the analysis of insulin secretion. The procedure is similar to our previous study with leucine [6].
Islet insulin output. Islets were isolated and incubated overnight, as described earlier. Then, the islets were incubated in 4-well plates (NUNC Brand Products, Roskilde, Denmark). Ten islets were incubated in 1 ml RPMI 1640 and 11.1 mM glucose each in the presence of 0.17, 1, 5, and 10 mM proline. Insulin output was defined as insulin concentration in the islet culture medium after the fixed incubation period. The incubation medium (25 μl) was removed after 24, 48, and 72 h, and stored at -20°C until insulin analysis. The procedure is similar to our previous study with leucine [6].
2.3 Cell culture
INS-1E cells (a generous gift from Prof. Claes B. Wollheim, Geneva, Switzerland) were cultured in RPMI 1640 medium containing 11.1 mM D-glucose at 37°C in a humidified atmosphere containing 95% air and 5% CO2. The medium was supplemented with:
- 10% FBS
- 100 IU/ml penicillin
- 100 μg/ml streptomycin
- 10 mM Hepes
- 2 mM L-glutamine
- 1 mM sodium pyruvate
- 50 μM 2-mercaptoethanol
The cells were passaged weekly.
Insulin secretion from INS-1E cells. The INS-1E cells were seeded (3.0 × 105 cells/well) onto 24-well Black Visiplate TC (Wallac Oy, Turku, Finland) plates in 1 ml RPMI 1640. After adhering overnight, the cells were cultured in RPMI 1640 with 11.1 mM glucose in the presence of 0.17, 1, 5, and 10 mM proline for 72 h in a humidified atmosphere (95% air and 5% CO2 at 37°C). Afterwards, the cells were preincubated with M-KRB, supplemented with 3.3 mM glucose and 0.1% human serum albumin for 30 min, and then the cells were incubated in 1 ml M-KRB containing 3.3 or 16.7 mM glucose for 1 h. Subsequently, 300 μl supernatant were collected and centrifuged, 200 μl of which were kept at -20°C for later insulin analysis. After the secretion study, the cells were washed once with 1 ml PBS (Gibco). Then, the number of cells was estimated using nuclear staining with SYTO 24 reagent (20 μl/well, Roche), and measured by FLUOstar Galaxy (BMG, Ramcon, Denmark). Unless otherwise stated, passage numbers between 60 and 82 were used.
Insulin output of INS-1E cells. INS-1E cells were incubated and treated as described earlier. For the insulin output study, the incubation medium (25 μl) was taken out after 24, 48, and 72 h, and stored at -20°C until insulin analysis. The procedure is similar to our previous study with leucine [6].
Insulin content of INS-1E cells. The INS-1E cells were incubated and treated as described above, but we used 6-well plates with a density of 1.0 × 106 cells/well in 3 ml RPMI 1640 instead of 4-well plates. After 72 h, the cells were washed once with 2 ml cold PBS (Gibco), and then 1 ml glycine-BSA buffer was added. Cells were scratched and sonicated twice at 0°C for 14 s (Branson Sonifier 250, Danbury, CT). We took 250 μl to evaluate the total protein content with a detergent-compatible protein kit (Bio-Rad Laboratories, Hercules, CA) for calibration of the insulin content. The remaining 750 μl were centrifuged for 30 min at 16,000 rpm, and the supernatant was collected and frozen at -20°C for a later insulin assay.
2.4 Insulin assay
Insulin was analyzed by radioimmunoassay using a guinea pig anti-porcine insulin antibody (Novo Nordisk, Bagsvaerd, Denmark), mono-125I-(Tyr A14)-labeled human insulin (Novo Nordisk A/S) as tracer, and rat insulin as standard (Novo Nordisk A/S). The separation of bound and free radioactivity was performed using ethanol. The inter- and intra-assay coefficients of variation were determined as <10%. The procedure is similar to our previous study with leucine [6].
2.5 Determination of triglyceride and cholesterol content
INS-1E cells were plated at 3.0 × 105 cells per well each in 24-well Black Visiplate TC plates combined with 1 ml of medium. The cells were allowed to adhere overnight. Then, they were incubated and treated as described earlier. After 72 h, the cells were washed once with 1 ml PBS, and the number of cells was estimated as described earlier. The medium was removed, and the cells were frozen for 1 h at -80°C. Subsequently, the cells were treated for 20 min with triglyceride (TG) reagents (250 μl/well, Roche) at room temperature. TG content was determined with a TG GPO-PAP kit (Roche), and normalized to cell number. The recovery of TG was ~90%.
Correspondingly, cells were treated for 20 min with cholesterol (CH) reagents (Roche; 250 μl/well) at room temperature. CH content was determined by a CHOD-PAP kit (Roche) and was normalized to cell number. The recovery of CH content was ~90%.
2.6 3H-thymidine incorporation in INS-1E cells
3H-thymidine incorporation was used to monitor INS-1E cell proliferation and DNA synthesis. Briefly, INS-1E cells (4 × 104/well) were seeded in 96-well isoplates (Wallac Oy, Turku, Finland), and cultured in RPMI 1640 with 11.1 mM glucose and 10% FBS. On the next day, the cells were incubated with "starvation medium" (RPMI1640 with 0.5 mM glucose, 0.1%BSA, without FBS) for 24 h. The cells were then cultured for another 72 h in RPMI 1640 (containing 0.5% BSA) and 11.1 mM glucose in the presence of 0.17, 1, 5, and 10 mM proline and 1 μCi (methyl-3H) thymidine (Perkin Elmer, Wellesley, MA, USA). After 72 h, the cells were washed twice in cold PBS and 200 μl liquid scintillator were applied. The incorporated (methyl-3H) thymidine was counted by a 1450 MicroBeta TRILUX (Wallac, Turku, Finland) device.
2.7 The incidence of dead INS-1E cells
INS-1E cells were seeded in 96-well Black Visiplate TC plates (Wallac Oy, Turku, Finland) at a density of 4 × 104 cells/well in 200 μl medium. The cells were allowed to adhere overnight. Then, they were treated and cultured with different concentrations of proline as described earlier. After 72 h, the number of dead cells in each well was calculated using a fluorometric assay kit based on the cell lysis and staining method [15] (Cytotoxic Fluoro-test Wako; Wako Pure Chemical Industries, Osaka) in the FLUOstar Galaxy. A linear relationship was confirmed in advance between the viable cell density and fluorescence intensity.
2.8 RNA extraction and cDNA synthesis
Three different passages of INS-1E cells were incubated in RPMI 1640 in the presence of 0.17 and 5 mM proline for 72 h. The RNA of treated cells was extracted using the AllPrep RNA/Protein kit (Qiagen, Valencia, CA, USA) on the QIAcube machine (Qiagen, Valencia, CA) according to the manufacturer's instructions (AllPrep RNA/Protein Handbook; Qiagen). RNA concentration and purity were determined by measuring the absorbance at 260 nm and 280 nm (NanoDrop ND-8000 UV-Vis Spectrophotometer, NanoDrop Technologies Wilmington, DE). The RNA quality was evaluated by examining the 18s and 28s ribosomal band on a 1% non-denaturing agarose gel, stained with SYBR green.
Reverse transcription (RT) was performed using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc. Hercules, CA, USA) according to the manufacturer’s instructions. We applied 1 μg total RNA for each 20 μl RT reaction.
2.9 Quantitative RT-PCR
RT-PCR was performed using an ABI 7500 FAST polymerase chain reaction machine (ABI; Foster City, CA). Predesigned TaqMan probes and primers with the following catalog numbers were obtained from Applied Biosystems (ABI, Foster City, CA, USA):
- Pancreatic and duodenal homeobox-l (Pdx1, Rn00755591_m1)
- v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MafA, Rn00845206_s1)
- Insulin1 (Ins1, Rn02121433_g1)
- Insulin2 (Ins2, Rn01774648_g1)
- Acetyl-CoA carboxylase α (ACC1, Rn00573474_m1)
- Fatty acid synthase (Fasn, Rn01463548_m1)
- Pyruvate dehydrogenase kinase 2 (Pdk2, Rn00578427_m1)
- Mevalonate (diphospho) decarboxylase (Mvd, Rn00579216_m1)
- Cytochrome c oxidase, subunit VIa, polypeptide 2 (Cox6a2, Rn00563091_g1)
- Caspase-3 (Casp3, Rn00563902_m1)
- Death-associated protein kinase 3 (Dapk3, Rn00574826_m1)
- Bcl2 modifying factor (Bmf, Rn00594968_m1)
- 18S (Hs99999901_s1)
The RT-PCR reaction mix contained:
- 5 µl of 2 × TaqMan FAST Universal Master Mix (P/N 43660783, ABI; Foster City, CA)
- 0.5 µl of 20 × TaqMan Assay/probe (ABI; Foster City, CA)
- cDNA equivalent to 50 ng total RNA in 4.5 µl H2O
The thermal FAST cycle program was one cycle at 95°C for 20 s, 40 cycles at 95°C for 3 s, and 60°C for 30 s. Each PCR sample was performed in six repeats, and gene expressions were normalized to eukaryotic18S expression. All assays were carried out in 96-well plates with an optical adhesive cover (P/N 4346906 and P/N 4311971 ABI; Foster City, CA). We used the 2-ΔΔCT method to calculate the relative gene expression. No template controls and no amplification controls were included as negative control for each gene.
2.10 INS-1E cell growth and lysis for proteomics
Proteins from three different passages of INS-1E cells were extracted at the same time as RNA was purified on the QIAcube machine. We extracted two samples at each passage and for each treatment condition, resulting in six replicates of each treatment, which were used for proteomics.
The protein content of the cell suspensions was analyzed with the Bradford assay (Bio-Rad). In order to obtain a sufficiently high protein concentration from the cell suspensions, the samples (250 µl) were precipitated under addition of 250 µl 24% tri-chloroacetic acid at 4°C, and left to precipitate for 30 min at 0°C, followed by centrifugation at 6000 g and 0°C for 10 min. The supernatant was removed, the protein pellet washed twice by addition of 500 µl acetone at 0°C, and centrifuged between each washing step, as described earlier. The protein pellets were kept at -80°C for proteomic analysis.
2.11 Proteomic analyses
The 2-dimensional gel electrophoresis (2-DGE) analysis was performed, as described earlier [16]. Briefly, the stored protein pellets were thawed and solubilized in 250 μl 2D-lysis buffer, and subsequently the tenfold amount of Millipore water was added. Based on protein determination using the Bradford assay, a sample volume corresponding to 100 µg proteins was applied to each gel. The 12 samples were each analyzed in a single 2-DGE gel set consisting of six gels representing either control INS-1E cells or cells treated with proline, as described earlier, using 11 cm IPG strips (pH 5-8) for the first dimension, and 12.5% Criterion gels (Bio-Rad) for the second dimension. Analytical gels were stained with Flamingo Pink (Bio-Rad, Hercules, CA, USA) and scanned using a Molecular Imager FX (Bio-Rad, Hercules, CA, USA). Images were analyzed with PDQuest (Bio-Rad, Hercules, CA, USA). Gels for spot identification were stained with colloid Coomassie Blue R-250 [17]. In-gel digestion for peptide mass fingerprinting, desalting, and concentration of protein spots, identification of MFGM proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry, and image analysis was performed, as described earlier [18].
2.12 Statistical analysis
We compared groups using one-way ANOVA. We also performed statistical analysis using the unpaired two-tailed Student's t-test with unequal variances. Each treatment condition was compared with controls using Bonferroni's correction. Differences of p < 0.05 were considered to be significant. Data are presented as mean ± standard error of means (SEM).
3. Results
3.1 Insulin secretion
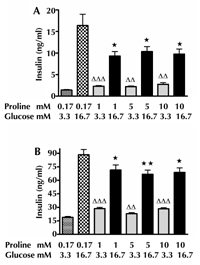
Figure 1A shows that at 3.3 mM glucose BIS from isolated islets increased after 72-h incubation with 1, 5, or 10 mM proline. By contrast, glucose (16.7 mM)-stimulated insulin secretion decreased after 72-h exposure of the islets to 1, 5, or 10 mM proline (Figure 1A). Similar results were obtained with INS-1E cells, demonstrating increased BIS and decreased GSIS after 72-h exposure to high proline concentration (Figure 1B).
 |
 |
Figure 1. Effects of 72-h incubation with proline on the response to glucose. The insulin secretion from mouse islets (A) at low (3.3 mM) (ΔΔ p < 0.01, ΔΔΔ p < 0.001) and high (16.7 mM) glucose concentrations (*p < 0.05) for 1 h. This part includes three independent experiments. For each experiment we applied the mean value obtained from 8-11 islets for each condition. Insulin secretion from 10,000 INS-1E cells (B) at low (3.3 mM) (ΔΔp<0.01, ΔΔΔp<0.001) and high (16.7 mM) glucose concentrations (*p < 0.05, ** p < 0.01) for 1 h. This part includes three independent experiments. For each experiment we applied the mean value obtained from six samples for each condition. |
|
3.2 The effect of proline on insulin output
Islets and INS-1E cells showed no significant differences in insulin output at 24, 48, and 72 h between 1-10 mM proline vs. control (data not shown), respectively.
3.3 Influence of proline on insulin content in INS-1E cells
No significant change in insulin content was observed after 72-h culture of INS-1E cells with 1, 5, or 10 mM proline vs. control (data not shown), respectively.
3.4 Effects of 72-h exposure to proline on gene expressions in INS-1E cells
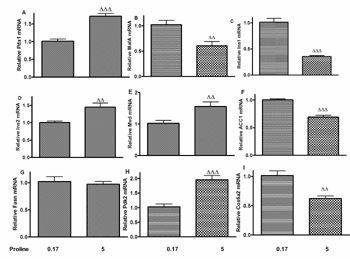
The impact of a 72-h incubation with 5 mM proline on gene expression was studied in INS-1E cells for a number of β-cell genes (Figure 2, A-I). Proline upregulated Pdx1 and Ins2 gene expression (Figure 2, A and D), and downregulated MafA (Figure 2B) and Ins1 gene expression (Figure 2C). As depicted in Figure 2E, Mvd mRNA expression was upregulated after proline treatment.
As illustrated in Figure 2, F-G, ACC1 mRNA level was significantly decreased (Figure 2F), while there was no change in Fasn gene expression (Figure 2G). As shown in Figure 2H, Pdk2 mRNA expression was increased, whereas proline reduced Cox6a2 mRNA level significantly in INS-1E cells (Figure 2I).
 |
 |
Figure 2. Changes in gene expressions in INS-1E cells induced by 5 mM proline. The following genes were investigated: Pdx1 (A), MafA (B), Ins1 (C), Ins2 (D), Mvd (E), ACC1 (F), Fasn (G); Pdk2 (H), Cox6a2 (I). ΔΔp < 0.01, ΔΔΔp < 0.001 versus control. RNA extraction includes three experiments applying different passage number cells. RT-PCR was performed in six repeats. |
|
3.5 Influence of proline on cholesterol and triglyceride content in INS-1E cells
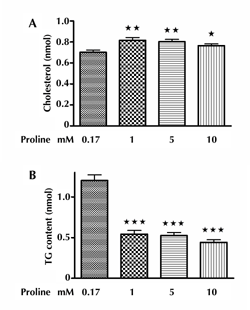
As can be seen in Figure 3A, the CH content in INS-1E cells increased by a 72-h culture of 1, 5, or 10 mM proline, respectively. Figure 3B demonstrated that the TG content of INS-1E cells exposed to proline (1, 5, or 10 mM) for 72 h was significantly decreased compared with that in controls.
 |
 |
Figure 3. Effect of proline on CH and TG content of INS-1E cells. Panel A shows the changes in CH content per 10,000 INS-1E cells after 72-h incubation with 1-10 mM proline, with * p < 0.05, ** p < 0.01 versus control. This part includes three independent experiments. For each experiment we applied the mean value from six samples for each condition. Panel B shows the changes in TG content per 10,000 INS-1E cells after 72-h incubation with proline (1-10 mM), with *** p < 0.001 versus control. This part includes three independent experiments. For each experiment we applied the mean value from six samples for each condition. |
|
3.6 Effect of proline on gene expression associated with apoptosis in INS-1E cells
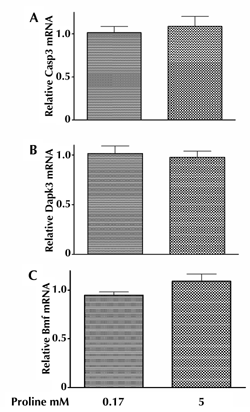
As shown in Figure 4, A-C, 5 mM proline did not affect mRNA levels of Casp3 (Figure 4A), Dapk3 (Figure 4B), and the pro-apoptotic protein Bmf (Figure 4C) in INS-1E cells.
 |
 |
Figure 4. Impact of proline on the expressions of genes associated with apoptosis. Alterations in mRNA expression in genes associated with apoptosis in INS-1E cells induced by 72 h exposure to 5 mM proline. Casp3 (A), Dapk3 (B), Bmf (C). RNA extraction includes three independent experiments. RT-PCR was performed in six repeats. |
|
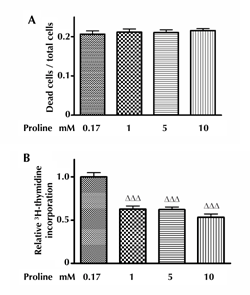
3.7 Impact of proline on the viability of INS-1E cells and 3H-thymidine incorporation in INS-1E cells culture
While the percentage of dead INS-1E cells was unchanged by 1, 5, and 10 mM proline (Figure 5A), proline (1, 5, and 10 mM) decreased 3H-thymidine incorporation in INS-1E cells, indicating a proline-induced suppression of INS-1E cell proliferation (Figure 5B).
 |
 |
Figure 5. Impact of proline on the viability of INS-1E cells and the incorporation of 3H-thymidine into INS-1E cells. Panel A shows the impact of 72-h incubation with proline (1-10 mM) on the viability of INS-1E cells. This part includes three independent experiments. For each experiment we applied the mean value from ten samples for each condition. Panel B shows the effect of 72 h incubation with proline (1-10 mM) on 3H-thymidine incorporation in INS-1E cells, with ΔΔΔp < 0.001 versus control. This part includes three independent experiments. For each experiment we applied the mean value from ten samples for each condition. |
|
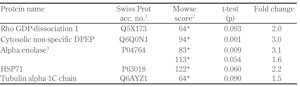
3.8 Proteomics: alteration in INS-1E cells induced by proline
Results from proteomic studies are presented in Table 1. Proline (5 mM) increased the amount of cytosolic non-specific dipeptidase in comparison with the control. Moreover, proline increased α enolase in two spots, at two different isoelectric point (pI) values, probably due to phosphorylation variants.
Table
1.
INS-1E cellular proteins identified by mass spectrometry and significantly affected by proline |
|
 |
 |
Legend:
1 Primary accession key in the SWISS-PROT database. 2 Score of the Mascot search. Significant scores (p < 0.05) are marked by *. 3 Alpha enolase was found to be differentially regulated in two spots, at two different pI values, probably due to phosphorylation variants. Abbreviations: DPEP – dipeptidase, GDP – guanosine diphosphate, HSP71 – heat shock protein 71. |
|
4. Discussion and conclusions
Elevated circulating levels of proline are found in subjects with IR, obesity, and T2D [5, 7]. The present study demonstrated that long-term exposure to proline caused β-cell dysfunction, with increased BIS and decreased GSIS in both isolated mouse islets and clonal rat INS-1E cells (Figure 1).
As shown in Figure 1 we have included controls that contain a proline concentration in the static incubation medium of 0.17 mM which reflects the normal circulating level in rodents. We applied similar exposure time (72 h) in controls and to the high doses of proline. The insulin responses to chronic exposure to proline are in line with our previous results with the AA, leucine, using the same doses and experimental conditions [6]. Thus, in response to chronic exposure both leucine and proline cause increased basal and impaired glucose-stimulated insulin secretion, indicating that an aminoacidotoxicity may operate like gluco- and lipotoxicity and contribute to impaired islet-cell function in T2D. It is however unknown whether the β-cell dysfunction in response to the two AAs has the same mechanisms of action.
It should be noted that we have added high amino acid concentrations to the incubation medium. These concentrations are similar to those used in other in vitro studies. The reason for this procedure is that the normal vascular supply is not present in isolated islet. We have assumed that higher AA concentrations are needed in the incubation medium to establish a milieu at the islet cell level that reflects supraphysiological circulating AA. It should however be noted that the effective levels of AAs within the isolated islets are unknown. In acute experiments, it has previously been demonstrated that both leucine and proline induce a stimulation of insulin secretion [11], but these results differ from long-term experiments.
Pdx1 and MafA are potent stimulators for the transcription of insulin genes [19]. In line with proline-induced β-cell dysfunction, proline decreased MafA mRNA expression in INS-1E cells (Figure 2B). This results in decreased Ins1 mRNA expression in proline-treated INS-1E cells (Figure 2C). It is well known that MafA binds to the C1 element within the insulin promoter. Mutagenesis of the human C1 MafA-binding site reduces insulin promoter activity by 74% in INS-1 β-cells [19]. In contrast to the downregulation of Ins1 mRNA expression, Ins2 mRNA expression increased significantly in proline-treated INS-1E cells (Figure 2D). This difference in the expression of Ins1 and Ins2 mRNA is puzzling. There was no change in total insulin content or output in proline-treated INS-1E cells. It has been proposed that the expression of the two insulin genes is independently regulated and coordinated in both rats and mice [20]. However, we have not found evidence that Ins2 is preferentially secreted during basal conditions [20]. In fact, Ins1 accounts for 60-90% of total insulin immunoreactivity in β-cell lines [20].
The increased Pdx1 mRNA expression found in this study is noteworthy (Figure 2A); it is in accordance with increased mRNA expression of Pdx1 found in isolated islets from T2D patients, despite declined insulin release in response to glucose compared with control islets [21]. It is also in agreement with our previous finding (with leucine) that Pdx1 gene expression is upregulated in INS-1E cells after long-term exposure to proline [6].
Accumulating evidence indicates that CH levels in β-cells play a pivotal role in the regulation of GSIS. We found that the cellular CH content of INS-1E cells increased after exposure to excess proline for 72 h (Figure 3A). This is in line with our microarray data [22], which demonstrated that incubation of 5 mM proline for 72 h downregulated 5' adenosine monophosphate-activated protein kinase (AMPK), but upregulated expressions of CH biosynthetic enzymes, e.g., Mvd, farnesyl diphosphate farnesyl transferase, and 7-dehydrocholesterol reductase in INS-1E cells [22].
As expected, proline increased Mvd mRNA expression (Figure 2E) in INS-1E cells, as proven by RT-PCR. Our observation of proline-induced AMPK downregulation is very interesting; it is in line with the reduced percentage of activated AMPK protein in isolated islets from T2D patients [21]. In contrast to increased CH content, we found a proline-induced reduction in the TG content (Figure 3B) in INS-1E cells, which may partly be due to proline-induced ACC1 downregulation (Figure 2F), while there was no alteration of Fasn mRNA (Figure 2G).
Conflicting evidence exists regarding the role of CH in insulin secretion. A decrease in insulin secretion was observed under long-term inhibition of CH synthesis in insulin-secreting cells [23]. The associated impairment of GSIS is probably due to an effect of reduction in the protein isoprenylation and more importantly through membrane disarrangement [23]. Thus, both increase [24-26] and reduction [23] in β-cell CH content have been shown to reduce insulin secretion. In conclusion, a critical concentration of cellular/membrane CH content is required for adequate secretion of insulin.
Metabolic profiling of β-cell function identified mitochondria as sensors and generators of metabolic signals controlling insulin secretion. Pyruvate dehydrogenase (PDH) may play a supportive role for GSIS by providing acetyl-CoA to form citrate for downstream shuttles. PDH activity is activated by PDH phosphatase and inactivated by Pdk through dephosphorylation and phosphorylation, respectively [27]. Proline-mediated upregulation of Pdk2 in INS-1E cells (Figure 2H) phosphorylates PDH and reduces its activity. Consequently, excess proline as in high FAs [28] and high glucose [28] reduces islet PDH activity, which may be involved in impaired GSIS by pancreatic β-cells.
It is well known that mitochondrial metabolism, which drives the respiratory chain and subsequently ATP production via oxidative phosphorylation (OxPhos), is required for normal GSIS in β-cell function. Our microarray data suggested that Cox6a2 gene expression was significantly decreased in proline-incubated INS-1E cells [22]. This was further validated in the present study using RT-PCR (Figure 2I). Interestingly, Weskler-Zangen et al. confirmed that the Cohen diabetic rat, which is a genetic model of nutritionally induced diabetes, could be linked directly to reduced OxPhos [29]. The critical regulatory role of ATP production by OxPhos is underscored by the observation that the cytochrome c oxidase (Cox) activity in islets from the transgenic MKR mouse (a dominant-negative IGF-1 receptor mutation specifically in skeletal muscle) was significantly reduced. As a consequence, the ATP/ADP ratio was decreased in response to high-glucose stimulation, followed by blocked GSIS, indicating reduced mitochondrial oxidative capacity [30]. Moreover, other studies are in accordance with the findings that reduced OxPhos and ATP content lead to impaired GSIS in β-cells of OPA-1-deficient mice [31] and islets of type 2 diabetic subjects [32]. This points to an important mechanism by which proline impairs GSIS in INS-1E cells, and which is supported by several lines of evidence from human diabetes. Decreased Cox activity has been demonstrated in islets from a diabetic patient [33], and plays a critical role in mitochondrial substrate oxidation in human β-cells with mutations in the mitochondrial genome [34].
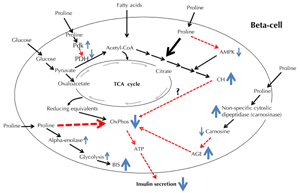
Interestingly, in CH-enriched β-cells, the glucose-mediated increase in cellular ATP content was dramatically reduced, and this was related to a decrease in glucose uptake via glucose transporter 2 and an impairment of mitochondrial metabolism [35]. Moreover, CH-enriched mouse β-cells have a low voltage-gated calcium channel density, which diminishes Ca2+ influx, and in turn, reduces insulin secretion [35]. The relationship between proline-induced CH accumulation and proline-induced Cox downregulation warrants further investigation (Figure 6).
 |
 |
Figure 6. The figure illustrates major pathways relating to the possible effects of proline in pancreatic β-cells. Solid-line arrows indicate a positive effect and dashed-line, red-colored arrows a negative effect. Size of arrows illustrates intensity of the effect. Abbreviations: AGE – advanced glycation end-products, AMPK – AMP-activated protein kinase, ATP – adenosine triphosphate, BIS – basic insulin secretion, CH – cholesterol, CoA – coenzyme A, OxPhos – oxidative phosphorylation, PDH – pyruvate dehydrogenase, Pdk – pyruvate dehydrogenase kinase, TCA – tricarboxylic acid. |
|
Proline-induced Cox6a2 mRNA downregulation may also be mediated through non-specific cytosolic dipeptidase. Previous studies on cytoplasmic dipeptide hydrolases of mammalian tissues have revealed a number of apparently distinct enzymes, including prolinase, carnosinase, and non-specific dipeptidase. Lenney proposed that the activity responsible for each of the three types of activity should be termed "non-specific cytosolic dipeptidase" [36]. Using proteomics, we found that non-specific cytosolic dipeptidase was increased in proline-treated INS-1E cells. Interestingly, AGEs impaired the GSIS of mice islets and INS-1 cells by inhibiting Cox activity and ATP synthesis [37]. Further studies are needed to clarify the probable association between non-specific cytosolic dipeptidase and AGE content in INS-1E cells exposed to excess proline (Figure 6).
Another protein identified as being differentially regulated was α enolase, which was identified in two spots, both of which were increased. Interestingly, Malmgren et al. confirmed that the rate of glycolysis was proportional to ambient glucose in the glucose-responsive 832/13 line [38]. In contrast, glycolysis was highly upregulated in the glucose-unresponsive 832/2 line, already reaching very high rates at a low glucose concentration, and exceeded that in 832/13 cells. These discrepancies could be explained by several alterations that affect glycolysis in 832/2 cells [38]. In accordance with these changes, higher long-term plasma glucose levels in islet donors correlated with upregulation of genes involved in glycolysis, whereas genes involved in mitochondrial metabolism were downregulated in human islets [38]. Consequently, elevated proline-induced α enolase in INS-1E cells could partly account for the increased BIS in INS-1E cells.
The amount of β-cell mass, at any given moment, is represented by the sum of replication, size, and neogenesis, minus the rate of apoptosis. Currently, there is no consistent agreement on the role of apoptosis in the pathogenesis of T2D. In the present study, proline had no effects on INS-1E cell death (Figure 5A). This finding is consistent with the observation that mice with a deletion of β-cell ABCA1 have increased CH content, but normal β-cell mass [26]. Collectively, increased CH content in β-cells does not mediate β-cell apoptosis. This was further supported by gene expression of key mediators in apoptosis using RT-PCR. There were no changes in the expression of Casp3, Dapk3, and Bmf genes in INS-1E cells (Figure 4) after 72 h excess proline exposure. Whereas, 3H thymidine incorporation experiments showed that INS-1E cell proliferation was reduced after 72 h exposure to elevated proline levels (Figure 5B). This suggests that elevated proline may cause reduced β-cell mass by decreasing β-cell mitotic division and proliferation.
In conclusion, exposure to excess proline causes increased BIS and impaired GSIS. These alterations are closely related to decreased insulin transcription and impaired mitochondrial OxPhos. Thus, aminoacidotoxicity may have detrimental effects on β-cell function, and may therefore play a pathogenic role in the development of T2D. However, further studies are necessary to validate our suggestions and hypothesis and to obtain a more complete picture of the mechanism exerted by elevated levels of AAs on islet cell function. As already stated, we have used very high AA doses in our incubation studies. Consequently, caution is required in interpreting the pathophysiological role of these doses.
Funding: This work was carried out as a part of the research program of the Danish Obesity Research Centre (DanORC, see www.danorc.dk). The study was supported by The China Scholarship Council, the Daloon Foundation, and the Institute of Experimental Clinical Research at Aarhus University.
Author contributions: ZL participated in the study design, carried out experiments and data analysis, and drafted the initial manuscript. LBL completed the proteomics experiment. PBJ, SG, and KH designed this project, and have read and approved the final manuscript.
Disclosures: The authors report no conflict of interests.
Acknowledgments:
We would like to thank Dorthe Rasmussen, Lene Trudsø, and Tove Skrumsager Hansen for skilled technical assistance and Michael Væth (Department of Biostatistics, Aarhus University) for statistical support.
References
- Hermansen K, Orskov H, Christensen SE. Streptozotocin diabetes: a glucoreceptor dysfunction affecting D cells as well as B and A cells. Diabetologia 1979. 17(6):385-389. [DOD] [CrossRef]
- Unger RH, Orci L. The essential role of glucagon in the pathogenesis of diabetes mellitus. Lancet 1975. 1:14-16. [DOD] [CrossRef]
- Xiao J, Gregersen S, Kruhoffer M, Pedersen SB, Orntoft TF, Hermansen K. The effect of chronic exposure to fatty acids on gene expression in clonal insulin-producing cells: studies using high density oligonucleotide microarray. Endocrinology 2001. 142:4777-4784. [DOD] [CrossRef]
- El-Assaad W, Joly E, Barbeau A, Sladek R, Buteau J, Maestre I, Pepin E, Zhao S, Iglesias J, Roche E, et al. Glucolipotoxicity alters lipid partitioning and causes mitochondrial dysfunction, cholesterol, and ceramide deposition and reactive oxygen species production in INS832/13 beta-cells. Endocrinology 2010. 151:3061-3073. [DOD] [CrossRef]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009. 9:311-326. [DOD] [CrossRef]
- Liu Z, Jeppesen PB, Gregersen S, Larsen LB, Hermansen K. Chronic exposure to leucine in vitro induces beta-cell dysfunction in INS-1E cells and mouse islets. J Endocrinol 2012. 215:79-88. [DOD] [CrossRef]
- Tai ES, Tan ML, Stevens RD, Low YL, Muehlbauer MJ, Goh DL, Ilkayeva OR, Wenner BR, Bain JR, Lee JJ, et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia 2010. 53:757-767. [DOD] [CrossRef]
- Tremblay F, Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem 2001. 276:38052-38060. [DOD]
- Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest 1998. 101:1519-1529. [DOD] [CrossRef]
- Takano A, Usui I, Haruta T, Kawahara J, Uno T, Iwata M, Kobayashi M. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol Cell Biol 2001. 21:5050-5062. [DOD] [CrossRef]
- Liu Z, Jeppesen PB, Gregersen S, Chen X, Hermansen K. Dose- and glucose-dependent effects of amino acids on insulin secretion from isolated mouse islets and clonal INS-1E beta-cells. Rev Diabet Stud 2008. 5:232-244. [DOD] [CrossRef]
- McClenaghan NH, Barnett CR, Flatt PR. Na+ cotransport by metabolizable and nonmetabolizable amino acids stimulates a glucose-regulated insulin-secretory response. Biochem Biophys Res Commun 1998. 249:299-303. [DOD] [CrossRef]
- Wu G, Bazer FW, Burghardt RC, Johnson GA, Kim SW, Knabe DA, Li P, Li X, McKnight JR, et al. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids 2011. 40:1053-1063. [DOD] [CrossRef]
- Lacy PE, Kostianovsky M. Method for the isolation of intact islets of Langerhans from the rat pancreas. Diabetes 1967. 16:35-39. [DOD] [CrossRef]
- Sakai K, Hayashi C, Yamaji H, Fukuda H. Use of nonionic surfactants for effective supply of phosphatidic acid in serum-free culture of Chinese hamster ovary cells. J Biosci Bioeng 2001. 92:256-261. [DOD] [CrossRef]
- Young JF, Larsen LB, Malmendal A, Nielsen NC, Straadt IK, Oksbjerg N, Bertram HC. Creatine-induced activation of antioxidative defence in myotube cultures revealed by explorative NMR-based metabonomics and proteomics. J Int Soc Sports Nutr 2010. 7:9. [DOD] [CrossRef]
- Kang DH, Gho YS, Suh MK, Kang CH. Highly sensitive and fast protein detection with coomassie brilliant blue in sodium dodecyl sulfatepolyacrylamide gel electrophoresis. Bull Kor Chem Soc 2002. 23:1511-1512. [DOD] [CrossRef]
- Bach Larsen L, Wedholm-Pallas A, Lindmark-Mansson H, Andren A. Different proteomic profiles of sweet whey and rennet casein obtained after preparation from raw versus heat treated skimmed milk. Dairy Sci Technol 2010. 90:641-656. [DOD] [CrossRef]
- Docherty HM, Hay CW, Ferguson LA, Barrow J, Durward E, Docherty K. Relative contribution of PDX-1, MafA and E47/beta2 to the regulation of the human insulin promoter. Biochem J 2005. 389:813-820. [DOD] [CrossRef]
- Linde S, Welinder BS, Nielsen JH. Analysis of proinsulin and its conversion products by reversed-phase high-performance liquid chromatography. J Chromatogr 1993. 614:185-204. [DOD] [CrossRef]
- Del Guerra S, Lupi R, Marselli L, Masini M, Bugliani M, Sbrana S, Torri S, Pollera M, Boggi U, Mosca F, et al. Functional and molecular defects of pancreatic islets in human type 2 diabetes. Diabetes 2005. 54:727-735. [DOD] [CrossRef]
- Liu Z, Luo Y, Jerpersen BP, Gregersen S, Hermansen K. Amino acid induced gene expression profiling in clonal betas cell line INS-1E cells. Diabetes Metab Res Rev 2011. 27:120-176. [DOD] [CrossRef]
- Zuniga-Hertz JP, Rebelato E, Kassan A, Khalifa AM, Ali SS, Patel HH, Abdulkader F. Distinct pathways of cholesterol biosynthesis impact on insulin secretion. J Endocrinol 2015. 224:75-84. [DOD] [CrossRef]
- Kruit JK, Wijesekara N, Westwell-Roper C, Vanmierlo T, de Haan W, Bhattacharjee A, Tang R, Wellington CL, LütJohann D, Johnson JD, et al. Loss of both ABCA1 and ABCG1 results in increased disturbances in islet sterol homeostasis, inflammation, and impaired beta-cell function. Diabetes 2012. 61:659-664. [DOD] [CrossRef]
- Peyot ML, Pepin E, Lamontagne J, Latour MG, Zarrouki B, Lussier R, Pineda M, Jetton TL, Madiraju SR, Joly E, et al. Beta cell failure in diet-induced obese mice stratified according to body weight gain: secretory dysfunction and altered islet lipid metabolism without steatosis or reduced beta cell mass. Diabetes 2010. 59:2178-2187. [DOD] [CrossRef]
- Brunham LR, Kruit JK, Pape TD, Timmins JM, Reuwer AQ, Vasanji Z, Marsh BJ, Rodrigues B, Johnson JD, Parks JS, et al. Beta-cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nat Med 2007. 13:340-347. [DOD] [CrossRef]
- Patel MS, Korotchkina LG. Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases. Exp Mol Med 2001. 33:191-197. [DOD] [CrossRef]
- Xu J, Han J, Epstein PN, Liu YQ. Regulation of PDK mRNA by high fatty acid and glucose in pancreatic islets. Biochem Biophys Res Commun 2006. 344:827-833. [DOD] [CrossRef]
- Weksler-Zangen S, Aharon-Hananel G, Mantzur C, Aouizerat T, Gurgul-Convey E, Raz I, Saada A. IL-1beta hampers glucose-stimulated insulin secretion in Cohen diabetic rat islets through mitochondrial cytochrome c oxidase inhibition by nitric oxide. Am J Physiol Endocrinol Metab 2014. 306:E648-E657. [DOD] [CrossRef]
- Lu H, Koshkin V, Allister EM, Gyulkhandanyan AV, Wheeler MB. Molecular and metabolic evidence for mitochondrial defects associated with beta-cell dysfunction in a mouse model of type 2 diabetes. Diabetes 2010. 59:448-459. [DOD] [CrossRef]
- Zhang Z, Wakabayashi N, Wakabayashi J, Tamura Y, Song WJ, Sereda S, Clerc P, Polster BM, Aja SM, Pletnikov MV, et al. The dynamin-related GTPase Opa1 is required for glucose-stimulated ATP production in pancreatic beta cells. Mol Biol Cell 2011. 22:2235-2245. [DOD] [CrossRef]
- Anello M, Lupi R, Spampinato D, Piro S, Masini M, Boggi U, Del Prato S, Rabuazzo AM, Purrello F, Marchetti P. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia 2005. 48:282-289. [DOD] [CrossRef]
- Kobayashi T, Nakanishi K, Nakase H, Kajio H, Okubo M, Murase T, Kosaka K. In situ characterization of islets in diabetes with a mitochondrial DNA mutation at nucleotide position 3243. Diabetes 1997. 46:1567-1571. [DOD] [CrossRef]
- Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L, et al. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet 2006. 79:54-61. [DOD] [CrossRef]
- Lee AK, Yeung-Yam-Wah V, Tse FW, Tse A. Cholesterol elevation impairs glucose-stimulated Ca2+ signaling in mouse pancreatic beta-cells. Endocrinology 2011. 152:3351-3361. [DOD] [CrossRef]
- Lenney JF. Human cytosolic carnosinase: evidence of identity with prolinase, a non-specific dipeptidase. Biol Chem Hoppe-Seyler 1990. 371:161-171. [DOD]
- Zhao Z, Zhao C, Zhang XH, Zheng F, Cai W, Vlassara H, Cairns NJ, Carter C, Cowley DJ, Duverger D, et al. Advanced glycation end-products inhibit glucose-stimulated insulin secretion through nitric oxide-dependent inhibition of cytochrome c oxidase and adenosine triphosphate synthesis. Endocrinology 2009. 150:2569-2576. [DOD] [CrossRef]
- Malmgren S, Nicholls DG, Taneera J, Bacos K, Koeck T, Tamaddon A, Wibom R, Groop L, Ling C, Mulder H, et al. Tight coupling between glucose and mitochondrial metabolism in clonal beta-cells is required for robust insulin secretion. J Biol Chem 2009. 284:32395-32404. [DOD] [CrossRef]
This article has been cited by other articles:
|