Diabetic Perspectives
| Rev Diabet Stud,
2005,
2(1):40-52 |
DOI 10.1900/RDS.2005.2.40 |
Genetic Susceptibility to Type 1 Diabetes in the Intracellular Pathway of Antigen Processing – A Subject Review and Cross-Study Comparison
Charles Sia1,  2 2
1Department of Immunology, United Biomedical Inc., 25 Davids Drive, Hauppage, New York 11788, USA.
2Department of Economics, University of Dortmund, 44227 Dortmund, Germany.
Address correspondence to: Charles Sia, e-mail: csia@unitedbiomedical.com
Keywords: type 1 diabetes, T cell receptor, antigen presentation, TAP, LMP
Abstract
Ligand binding grooves of MHC class I molecules are able to load a panel of endogenous peptides of varying length and sequence derived from self or foreign origin to activate or deactivate cytotoxic CD8+ T cells. Peptides are assembled with class I molecules by pathways that are either dependent or independent of transport by ABC proteins (TAP) and degradation in the immunoproteasome by its subunits LMP2 and LMP7. Those peptides that require TAP and LMP treatment appear to be subject to control and optimization by TAP for proper customizing and efficient presentation. Therefore, allelic variations in the coding sequences of TAP and LMP were suspected for a long time to be responsible for improper antigen processing, interruption of self-peptide presentation and reduced cell surface expression of MHC class I molecules resulting in the activation of autoreactive CD8+ T cells. In this article we reviewed the controversial findings regarding the role of TAP and LMP genes in autoimmune diabetes and reevaluated data of eleven separate studies in a cross-study analysis by genotype and HLA haplotype matching. We could confirm previous results by showing that TAP2*651-A/F and TAP2*687-A/A are significantly associated with disease, independently of linkage disequilibrium (LD). LMP2-R/H surprisingly seems to be primarily disease-conferring although a weak association with DR4 serotypes can be observed. Our analysis also suggests that LMP7-B/B, TAP1-A/A and TAP2*687-A/B are the protective genotypes and that these associations are not secondary to LD with DRB1. Consequently, intracellular antigen processing associated with TAP- and proteasome-dependent pathways seems to be a critical element in T cell selection for the retention of a balanced immunity.
Introduction
Major histocompatibility complex (MHC) class I molecules are expressed on any human cells except erythrocytes and trophoblasts. These molecules are essential for the presentation of endogenous peptides that can be of the body’s own or of foreign (viral) origin on the surface of virus-infected or antigen-presenting cells (APC) [1]. Usually, CD8+ cytotoxic T lymphocytes (CTL) can recognize peptides presented at these sites of cells using a receptor consisting of an α- and a β-chain together with the CD3 complex, which is known as the T cell receptor (TCR), and form an adherent binding site using their CD8 molecule (Figure 1) [2, 3]. In recent times it has been shown that even alloreactive CD4+ T cells, which are assumed to play a major role in the preclinical period of autoimmune diabetes, are capable of recognizing self-peptides loaded on MHC class I epitopes [4-6]. The recognition of virus-induced self-peptides on cell surfaces by an autoreactive class I T cell repertoire is considered to be a critical impetus for the establishment of the autoimmune response against pancreatic β-cells and the onset of clinical disease [6, 7]. The mechanism is still not understood in detail. Basically, MHC class I molecules are able to present a large amount of different peptides [8]. The class of peptides presented by these molecules determines the repertoire of specificities exhibited [9]. TCRαβ seem to have an inherent structural capability to react with diverse peptide/MHC structural patterns even other than the original patterns that might have been used to positively select that TCR [8]. Furthermore, the set of TCRαβ specific for a definite peptide/class I pattern may be highly heterogeneous [10]. This may allow for a high incidence of cross-reactivity which seems to reflect a feature of the positive selection mechanism in the thymus and the need for T cells in the repertoire to have an expanded capability for responding to a large variety of foreign antigens [8].
 |
 |
Figure 1. Recognition of an antigen presenting cell (large) by a CD8+ T cell (small) via interaction between TCR and MHC class I antigen epitope. |
|
In normal environments antigen specificity and self-reactivity is controlled by regulatory factors in the APC/T cell interaction. This is achieved by the usage of co-receptors and the induction of apoptotic programs to maintain a proper and balanced T cell selection [11, 12]. Upon appearance of failures in the antigen-processing pathway, proper T cell selection may be impaired resulting in the induction of autoreactive T cells [13]. In order to prevent the subsequent lymphocyte-mediated destruction of pancreatic β-cells or to reverse autoimmunity of this kind it is thus necessary to understand the genetic determinants of functional mechanisms involved in the MHC class I-induced activation of site-specific autoreactive T cells.
Peptides derived from exogenous sources such as bacteria are presented by MHC class II molecules on the surface of APC. The intra-cellular antigen processing is different from that of endogenous peptides in that these proteins are synthesized in the cytoplasm and the assembly of the peptide/class II molecule in the ER is augmented by the invariant chain. While MHC class I molecules are encoded by human leukocyte antigen (HLA)-A, -B, and -C, MHC class II is encoded by HLA-D genes. Many studies have reported evidence for the participation of both class I and class II genes in the pathogenesis of T1DM. Much emphasis has centered on DR and DQ and their associated haplotypes that may have a positive or a negative association with T1DM [14-16]. In comparison, there is information available on the association of class I genes with diabetes is smaller than those for class II genes. Possibly, the association of class I genes and T1DM was not apparent since variations in these genes, if involved in diabetic etiology, should lead to the same defects in all human cells and impaired antigen presentation may induce survival of potential target cells, as observed classically in carcinoma cells [17], rather than their destruction. The mechanism, therefore, may be attributed to the observation that, under consideration of factors responsible for specific antigen processing, cellular autoimmune responses mediated via class I molecules can indeed be tissue-specific caused by phenomena termed cross-reactivity of antigenic presentation to CD8+ T cells, which were first observed in transplantation studies [18, 19]. This article, therefore, follows the notion that T1DM may be hallmarked by a defective immune response resulting from failures in the presentation of self-peptides and thus focuses on the possible involvement of antigen-processing genes.
Antigen processing in the class I pathway of epitope presentation and its genetic basis
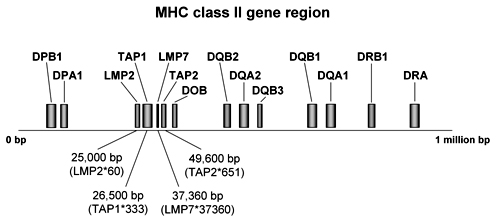
Endogenous peptides are generated by proteolytic degradation of endogenous peptides mediated by the proteasome and translocated from the cytosol into the lumen of the ER by transporters associated with antigen processing (TAP) for assembly with HLA class I molecules [20, 21]. After transport into the Golgi apparatus, compounded HLA class I/antigen complexes are presented on cell surfaces of APC forming the epitope for interaction with CD8+ T cells. The TAP peptide transporters belong to the adenosine triphosphate (ATP)-binding cassette transporters that use energy provided by ATP to translocate solutes across cellular membranes and, besides antigen presentation, they are involved in diverse processes, such as signal transduction, protein secretion, bacterial pathogenesis and sporulation [22, 23]. TAP molecules are heterodimers encoded by the TAP1 and TAP2 genes which are located within the MHC class II gene region between the immune-responsible DP and DQ genes at several codons in a region of 25-50 kb in length (Figure 2). TAP1 codes for the hydrophobic transmembrane and TAP2 for the carboxyl-terminal nucleotide-binding domain of the transporters [24] with the latter domain energizing the peptide transport. A peptide binds to a site shared between TAP1 and TAP2 and this has been mapped to the cytoplasmic loops in the membrane domains of the subunits [25].
 |
 |
Figure 2. MHC class II gene region. The map shows genes which are relevant in the etiology of T1DM. Positions of TAP and LMP genes were determined in several studies by amplification refractory mutation system (ARMS)-polymerase chain reaction (PCR) [41, 72]. TAP and LMP genes are centromeric in the MHC class II region to DPA1 and DOB. |
|
TAP is a critical element in the antigen processing pathway. This is shown by its selective transport of peptides with appropriate affinity for binding to class I molecules capable of inducing CTL responses [26, 27]. Peptides may also be associated with class I molecules via cytosolic processing pathways independently of active transport by ATP-binding cassette (ABC) proteins into the ER if they are expressed carboxyl-terminal to an ER signal sequence derived from an adenovirus glycoprotein [28, 29], but those that become associated with the empty Iα chain in the ER are required to be transported by TAP into this compartment. Newly synthesized empty class Iα chains bind to the multi-component loading complex consisting of TAP, Tapsin and Calreticulin allowing only the release of class I molecules with stably bound peptide ligands, which affirms the role of TAP to function as an intracellular checkpoint [30]. It was successfully shown that mutant HLA molecules that load peptides which are rapidly released from the cytosol to the cell surface in a TAP-independent manner appear to be instable [31]. Much evidence for the relevance of TAP for intracellular peptide kinetics and proper antigen presentation has been gathered by experiments with TAP knockout cells or modified TAP genes. Thus antigen presentation may be completely blocked if a cell is TAP-deficient [32] and the mechanism of peptide assembly and intracellular transport is impaired as a result of non-functional TAP genes [33]. Furthermore, defective TAP subunits give rise to reduced MHC class I expression and render APC to become ineffective in inducing CD8+ T cells [34]. This phenomena have been suggested to induce the positive selection of self-reactive CTL [35] and correlate with progression to diabetic autoimmunity in NOD mice and humans [36]. Interestingly, NOD mice carrying the rare TAP1B allele are reported to express conformationally abnormal class I molecules on their cell surfaces inducing the emergence of lymphocytes resistant to lysis by class I restricted CTL [37]. This has guided the attention to the existence of polymorphisms in TAP coding sequences associated with diabetic pathogenesis. Several randomized controlled studies were carried out since the early nineties, but the results have appeared to be controversial. Some results does not support the relevence of TAP genes in autoimmune diabetes [38, 39]. Others found significant associations, in particular with TAP2*687 and TAP2*651 [40, 41]. It thus remains questionable which role TAP may capture in the pathogenesis of T1DM. These uncertainties accentuate the need for further investigations.
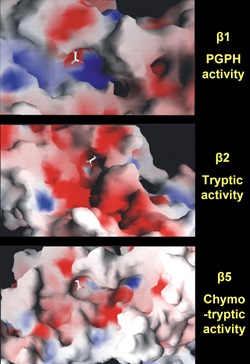
Another group of molecules critical for antigen processing includes the large multifunctional proteases, LMP2 and LMP7, which are also involved in the pathway of epitope delivery by degradation of proteins, protein cleavage, and catalysis of antigen processing generated in the proteasome. After endocytosis and intermediate storage in the endosome, where peptides are acidificated and translocated into the cytosol, antigens are fragmented and trimmed in the proteasome prior to further processing in the ER. LMP molecules are subunits of the 20S proteasome that are responsible for processing antigens with high into those with low molecular weight. They particularly modifies peptidase activities to facilitate the production of those types of peptides that are preferentially transported into the ER and selectively bound to MHC class I molecules. The 20S proteasome has two subunits (α and β) with several active sites in the seven β-subunits. Crystal structure imaging revealed that the β-subunits can cleave peptides after hydrophobic (chymotrypsin-like activity in β5), basic (trypsin-like activity in β2) and acidic (peptidylglutamyl peptide-hydrolyzing (PGPH) activity in β1) residues (Figure 3) [42, 43]. Under the influence of IFN-γ, the 20S proteasome is converetd into the immunoproteasome with its subunits being replaced by LMP2, LMP7 and multicatalytic-endo-peptidase-complex-like-1. The proteasome cleaves proteins to generate the exact C-terminal of CTL epitopes, and the N-terminal with possible extension of three amino acids or more. N-terminal trimming thus plays an important role in antigen presentation since the peptides that are loaded onto MHC class I ligand binding grooves are usually not more than eight to ten amino acids long [44]. The proteases increase the rates of cleavage after basic and hydrophobic residues and decrease cleavage after acidic residues [45]. The corresponding coding sequences are closely located to the TAP genes between the DP and DQ loci in the MHC class II gene region (Figure 2). The proximity of LMP and TAP genes to the highly diabetes-susceptible HLA region have cast them into doubt of linkage disequilibrium (LD).
 |
 |
Figure 3. Crystal structures of the proteasome 20S surface. The images represent the crystal structures of 20S proteasome surfaces. Cleavage of peptides takes place at active sites of the β-subunits after acidic (peptidylglutamyl peptide hydrolyzing (PGPH) activity in β1), basic (trypsin-like activity in β2), and hydrophobic (chymotrypsin-like activity in β5) residues. Each image shows the nucleophilic Th1 in sticks, the basic residues in blue, the acidic residues in red, and the hydrophobic residues in white. Image courtesy by Olivier Coux, CRBM-CNRS, France. |
|
The contributing role of the proteasome in generating CTL has been gathered through the observation that this proteolytic complex indeed degrades ubiquitinized proteins from the CTL-restricted parent proteins [46, 47]. The lack of one subunit leads to reduced levels of MHC class I cell-surface expression and to inefficient presentation of endogenous antigens [48]. Alterations in LMP genes are considered to be responsible for impaired antigen processing and reduced expression of class I/peptide complexes [49], which may result in deficient education and positive selection of self-specific CTL. Allelic variants of LMP genes in inbred mice are reported to be responsible for differences in the selection of peptides presented on the cell surface of APC giving rise to abnormal immune responses [50]. However, other lines of experimental data suggest that the proteasome plays a less significant role in the production of peptides to associate the different polymorphic forms of the MHC class I molecules. In LMP2 and LMP7 knockout mice and cells that lack either or both of these subunits, normal presentation of viral and intracellular derived peptides on the cell surface has been observed [51, 52]. Studies analyzing the relevance of LMP genes in T1DM in different populations as a result show divergent outcomes [38, 53, 54]. The information collected from the various studies suggests that proteasome-dependent and proteasome-independent mechanisms may act in intracellular kinetic pathways and in the generation of the spectrum of class I binding peptides. This observation resembles the findings that have been derived from studies on the association of TAP genes with T1DM and has led to the consideration that the relevance of both compartments in autoimmune disease is still an open question and requires further examination.
The mechanisms of how conformational changes in the primary sequences of LMP and TAP molecules contribute to peptide formation are not yet fully understood. How can we derive their relevance in the pathogenesis of autoimmunity? Generally, the process of intrathymic T cell selection is associated with the presentation of MHC/peptide complexes on APC to premature thymic T cells, and the level of this interaction reflects the degree of positive and negative selection of T cells [55]. The repertoire of receptors with which mature T cells are featured is shaped by interactions of this kind and by the number of TCRαβ engaged with peptide/class I complexes. Thereby, low numbers have been observed to correlate with positive and high numbers with negative selection [56]. The dimension of the TCRαβ repertoire thus depends on the diversity of intrathymic epitope supply, which appears to be closely associated with the pathway of antigen/class I expression [57]. Polymorphisms in LMP and TAP genes may account for a limited and variable expression of islet antigens in the thymus, contributing to the loss of self-tolerance against pancreatic β-cells [58]. The emergence of variant or uncommon polymorphic molecules in the MHC class I pathway of epitope presentation may thus critically influence susceptibility to autoimmune and other diseases [59].
The efficiency of peptide transport into the ER has been suggested as being dependent on the peptide-binding ability of the individual's TAP subunits [60]. Structural differences in either TAP1 and TAP2 variants induced by genetic polymorphism may result in a loss of binding ability or in a change of binding specificity thus influencing antigen presentation for the activation of CD8+ effector T cells. The endogenous peptide processing seems to be indeed allele-specific as different subsets of peptides bind to different antigen-binding grooves of MHC molecules under specific genetic variation [61, 62]. As the absence or deficiency of TAP can result in low class I surface expression this may provoke positive selection of self-reactive T cell repertoires [13] and confer the emergence of autoimmune diabetes finally [36]. Decreased peripheral interaction of T cells and self-peptide/class I molecules seems to be linked with this phenomenon [63]. Thus differences in binding selectivity appear to be a determinant for the observed alterations in immune responses, and this may be associated with genetic differences in the pathway of intracellular peptide processing. Therefore, allelic variations of genes that are responsible for intracellular transport kinetics in concert with other genes involved in the class I pathway of epitope delivery may lead to aggravation in antigen processing and thus may be considered as being disease conferring in the pathogenesis of T1DM. These considerations together with the "strategic" position of LMP and TAP genes in the HLA region has prompted investigators to test for the association of these genes with T1DM and whether they are in LD with HLA genes.
Association of LMP and TAP genes with T1DM
The presence of dimorphic sites on TAP genes led to the identification of 5 TAP1 (TAP1-A to TAP1-E) and 8 TAP2 (TAP2-A to TAP2-H) allelic variants [40, 64]. A and B are the most common alleles for instance coding for amino acids Ile and Val (Stop and Gln) at position TAP1*333 (TAP2*687 respectively). Other alleles have either been observed at very low frequency or not at all. Early studies have reported that the homozygous genotype TAP2-A/A, defined by sequence analysis that identified dimorphic patterns at position 687, exists at relatively high frequency in an Italian population of T1DM patients, whereas the TAP2-B allele was observed in a negative relation to T1DM [65]. It is, however, disputed if the observed associations are primary to the disease or secondary to LD between TAP2 and DQB1 or DRB1. According to Caillat-Zucman et al. [66] the negative association is independent of LD, whereas others found that the positive association is due to strict LD between DR and TAP2 [54, 67]. TAP1 alleles, on the other hand, seemed to have little or no association with diabetes in the Italian as well as in other populations [54, 65, 66, 68].
Allele TAP2-F was first analyzed in a large population of Northern American T1 diabetics of Caucasian origin by Jackson and colleagues in 1995 [40]. Due to the conformity of this allele to the frequently ocurring TAP2-A allele at all known positions except at position 651 causing amino acid substitution of Arg to Cys and to the fact that alleles had not always been tracked by full-length cDNA analysis until that time, it is reasonable that previous studies counted TAP2-F as TAP2-A. Jackson et al. tracked a polymerase chain reaction (PCR) product of 2191 bp that included the entire translated portion of the cDNA and some untranslated regions. They found that TAP2-F much more frequently exists in patients than in healthy controls [40]. Homozygosity of this allele was not detected in the 208 controls and heterozygosity with A was encountered in patients almost four times more frequently than in controls (52:14).
Most of the LMP2 and LMP7 alleles analyzed in randomized controlled studies carried out between 1994 and 1997 were observed no more frequently in patients than in controls [39, 53, 54, 69]. Two LMP2 alleles at position 60 causing an Arg to His substitution were widely analyzed, generally identified by sequence-specific oligonucleotide typing. In a study by van Endert and co-workers, LMP2-R homozygosity appeared twice as frequently in healthy individuals, suggesting that this allele has a protective property [54]. Subsequent studies reported that this is due to LMP7-R/R being in LD to DRB1*04-DQB1*03 [53, 69]. The results of studies analyzing the association between LMP7 alleles at position 37360 and T1DM are contradictory. While Undlien et al. found a significant association of genotype LMP7-A/B with healthy individuals [69], Deng and colleagues detected a higher frequency of this type in American T1DM patients of Caucasian origin (103:83) [53]. In contrast, homozygosity of the A allele was increased in patients (p = 0.02) and B homo-zygosity was found three times as frequently in healthy subjects than in patients (p < 0.001) in the Deng study [53]. Both effects were found to be independent of LD with HLA-DR or -DQ haplotypes. In a younger study, the protective property of the LMP7-B/B effect was confirmed [70].
The heterogeneity of results regarding the association of LMP and TAP genes with T1DM raises the question of how these differences could have originated. One possible explanation is that most of the study samples are too small to yield statistical robustness. Statistical tests are also rather sensitive to the process of data collection and identification. Early studies of TAP genes applied sequence-specific oligonucleotide analysis which hazards the consequence of ignoring similar but different alleles that are included in the full-length of the DNA. A study that analyzes a larger population could clear some of the statistical considerations and balance biases in data collection and identification. For this reason we carried out a cross-study analysis comprising data collected from eleven separate studies in the following section.
Cross-study analysis of LMP and TAP gene susceptibility to T1DM
Data and methods
We have compared studies that investigated the relevance of genes involved in the pathway of antigen processing (LMP and TAP) for susceptibility to T1DM in human patients. The general parameters of the studies incorporated in our cross-over comparison are summarized in Table 1. In respect to LMP2, five studies (van Endert et al. [54], Deng et al. [53], Chauffert et al. [39], Undlien et al. [69], Ding et al. 2000 [71]) were compared that typed intronic polymorphisms at position 60 (causing amino acid substitution Arg to His) containing a total of 616 T1DM patients and 709 controls (616/709), 441 of which are DR-matched. Considered data of studies regarding other genes are:
- LMP7 (position 37360, codons for Val/Leu substitution, 3 studies (Deng et al. [53], Undlien et al. [69], Ding et al. 2001 [70]), 460/499 subjects),
- TAP1 (position 333, codons for Val/Ile substitution, 5 studies (Colonna et al. [65], Kawaguchi et al. [38], van Endert et al. [54], Chauffert et al. [39], Ma et al. [72]), 364/371 subjects),
- TAP2 (position 687, codons for Gln/Stop substitution, 4 studies (Colonna et al. [65], van Endert et al. [54], Jackson et al. [40], Chauffert et al. [39]), 291/273 subjects),
- TAP2 (position 651, codons for Arg/Cis substitution, 2 studies (Jackson et al. [40], Penfornis et al. [41]), 299/158 subjects).
Table
1.
General parameters and reported results of studies included in the cross-over comparison |
|
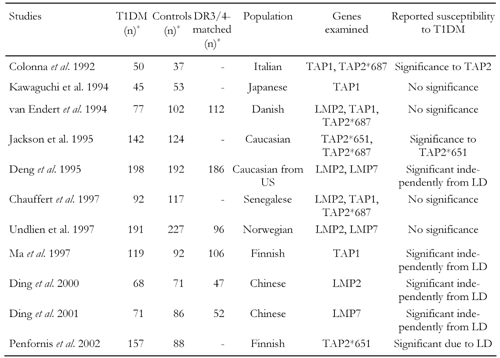 |
 |
Legend:
n: number of subjects, DR3/4-matched: number of subjects with identical haplotypes either DR3 or DR4, LD: linkage disequilibrium. * Maximum numbers included in the cross-over study (numbers can be smaller for different genotypes). |
|
Altogether 4040 genotypes (2030 T1DM patients and 2010 controls) collected from 2399 subjects (1210 T1DM patients and 1189 controls) from 11 separate studies have been included in our cross-over study (Table 2). Note that Table 1 indicates the numbers of subjects included in our study, but some of them are genotyped for more than one gene. Table 2 shows the numbers of subjects specified per gene. Therefore, total numbers of subjects in Tables 1 and 2 cannot correspond, since data from some individuals are used several times for analysis.
Table
2.
Genotype matching, studies, and total numbers of subjects per gene |
|
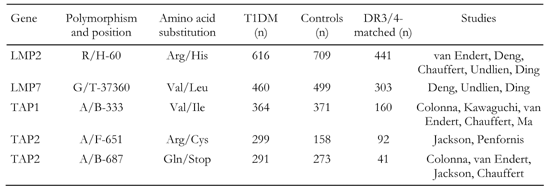 |
 |
Data were compared using χ2 test without Yates' corrections to determine cross-over significance. Bonferroni corrections for p-values were used where appropriate. LMP and TAP alleles as well as DR3/4 haplotypes were used as matching criteria for the eleven data sets.
Polymorphisms in T1DM patients
Studies analyzing the association of TAP and LMP alleles with T1DM draw different pictures. We have first compared studies and examined whether the frequency of genotype appearance is independent from group membership. Cross-over data demonstrate significant dependencies in genotype frequencies and group membership in all analyzed genes (Table 3). Polymorphisms in TAP and LMP genes seem to be significantly associated with disease (we take a closer view on the transmission of risk and protection mediated by specific genotypes in the subsequent section). Table 3 shows that, among the disease conferring TAP genotypes, TAP2*651 (p=0.005) and TAP2 *687 (p=0.001) are most strongly associated with diabetes. Apparently, genotypes B/B and F/F are rare variants of the TAP genes. The study samples do not seem to be large enough to find numbers of individuals bearing these types sufficiently often for imposing statistical power. As their number is small we face the hazard that the obtained specifications and identified distributions of B/B and F/F genotypes remain non-representative.
Table
3.
Independence test of LMP and TAP genes obtained from cross-over data |
|
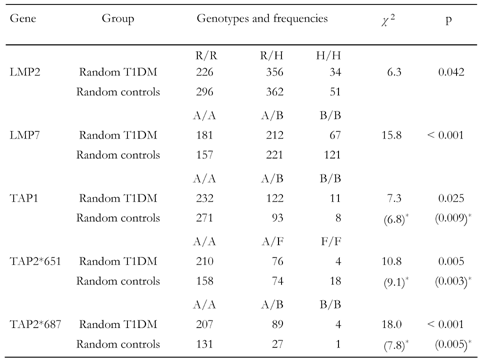 |
 |
Legend:
* Genotype B/B (F/F) omitted from calculation. |
|
Moreover, small differences in the distribution of these types may have a strong impact on the statistical significance and can result in an excessive distortion. Therefore, we have also calculated the χ2- and p-values for the examination of TAP gene susceptibility to T1DM adjusted by the omission of these types (parenthesized values in Table 3). Significance of susceptibility to T1DM rises for TAP1 and TAP2*651 and decreases slightly for TAP2*687 by this procedure, whilst remaining far below the marginal value of p = 0.05 for accepting dependency, suggesting that TAP genes are disease-conferring candidates. Apparently, the frequency is by far higher for all TAP2 genotypes in diabetic patients, which makes these genes susceptible.
It is also striking to observe that only one separate study shows significantly higher frequencies of allelic variations linked to healthy subjects, namely Undlien et al. for LMP7 (Table 4). Data from other studies either show no significance (van Endert et al. [54], Chauffert et al. [39], Undlien et al. [69]), or that polymorphism of different allele variation is associated with disease (Deng et al. [53], Ding et al. 2000 [71], Ding et al. 2001 [70]). This phenomenon may be attributed to the sample sizes analyzed in the separate studies or to biases in sample selections. In this regard, it is obvious that the frequencies of LMP7-A/A and LMP7-B/B genotypes presented in Ding et al. 2001 [70] show the inverse relation to the study by Undlien and colleagues [69]. B/B is the most frequent genotype in the study by Ding and colleagues from 2001 (A/A:B/B = 5:28 in patients and 9:50 in controls, as compared to 107:13 and 107:9 in the study by Undlien et al.). Should the types be inverted in one of the studies? We have taken the data as they appeared in the articles. The example corroborates, however, that statistical tests of hypotheses are rather sensitive to timing of data collection and recording procedures. Such effects are balanced by cross-over data pooling and are, therefore, largely eliminated in our analysis.
Table
4.
Polymorphisms of TAP and LMP in non-DR-matched subjects obtained from separate studies |
|
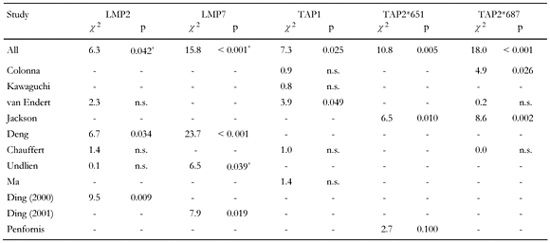 |
 |
Legend:
Values calculated as frequencies of genotype differences in T1DM vs. controls. * Genotype frequencies higher in controls. |
|
Association of genotypes to disease
In this section we address the question of whether particular genotypes can be associated with disease. The computation of the global test statistic χ2 is not sufficient to explain detailed dependencies. We need continuative information about the model adaptation which may be obtained by the examination of single components. Table 5, which contains uncorrected p-values (to correct multiply by three), shows the standardized residues demonstrating that there are large differences between frequencies observed and those expected in consideration of independence. Values above 1 or below -1 mean that the observed values may differ significantly from those which must actually have been expected. Negative (positive) values mean that the distortion is directed downwards (upwards). We are now able to easily read off from Table 5 to find that genotypes LMP2-R/H, LMP7-A/A, TAP1-A/B, TAP2*651-A/F and TAP2*687-A/A are strongly associated with disease. All significance levels of these genotypes are close to p = 0.01 or far below. Even after correction, p-values remain significant. In contrast, LMP2-R/R, LMP7-B/B, TAP1-A/A, TAP2*651-A/A and TAP2*687-A/B seem to be the protective genotypes. Their frequency is significantly smaller in patients. TAP2*651-F/F and TAP2*687-B/B are the rare types and cannot, therefore, be considered as representative. The result suggests that polymorphisms in LMP and TAP genes have a significant influence on disease onset or protection from disease.
Table
5.
Standardized residues and significance levels of genotypic associations with T1DM |
|
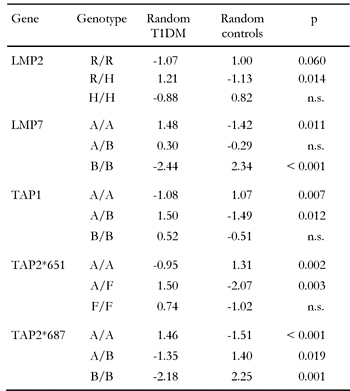 |
 |
In this light, the contrary character of results from the separate studies that divides them into two camps endorses the existence of biasing effects. The studies of the two camps found converse frequencies of LMP2-R/R and LMP2-R/H as well as LMP7-A/A and LMP7-A/B in the subject groups. LMP2-R/R is the protective genotype in Ding et al. 2000 [71] and Deng et al. [53], but is less frequent (and without significance) in healthy controls than in diabetics in the studies by Chauffert et al. [39], van Endert et al. [54] and Undlien et al. [69]. The opposite applies for LMP2-R/H, which is predisposing in Ding et al. and Deng et al. but more frequently existing in control subjects (although without significance) in the studies by Chauffert et al. [39], van Endert et al. [54] and Undlien et al. [69]. A similar logic applies for LMP7. Type B/B is the protective one in our study, which corresponds to Deng et al. [53] and Ding et al. 2001 [70], but this type appeared less frequently in the study by Undlien et al. [69].
Data from almost all separate studies that tested for TAP1 susceptibility show no significant association to both groups diabetics and controls (Table 4). Cross-over analysis, however, reveals an association of TAP1-A/B to disease and an unexpected small appearance of TAP1-A/A in diabetic patients, which accounts for a strong relationship, in particular as we would usually expect that by pooling data results derived from smaller samples would be diluted and equalized instead of aggravated. Although the frequencies for single genotypes in the separate studies differ in a contrary fashion without showing significance, the reason for this phenomenon may be attributed to the fact that a strict relationship exists which is only revealed by discarding biases of data collection.
Examination of linkage disequilibrium between LMP/TAP and DR genes
To investigate the influence of LD with HLA to LMP and TAP genes, we need to match subjects regarding their HLA genotypes. However, genotyping in the separate studies is not consistent and is afflicted with varying degrees of imprecision. For example, van Endert et al. only typed DR3/4 serotypes without identifying genotypes or corresponding alleles for determining homozygosity, which the authors attributed to unsuccessful PCR amplification [54]. Chauffert et al. determined several DQA1 and DQB1 alleles and combined them to yield indefinite genotypes in the fashion of DQB1*0201/x [39]. Thus, most of the studies included in our analysis have identified only a few or imprecise haplotypes. Considering that genotype-matching is incomplete and there are a plenty of other non-matched haplotypes, we do not certainly know which relation of HLA-genes to allelic differences between patients and controls are critical or definitely based on LD. In this light, carrying out a HLA-matching analysis may remain imprecise in any case and the results can only be tendentious.
In order to carry out an LD analysis we had to choose a common haplotype under the restriction of (1) avoiding a high degree of imprecision and (2) allowing for the selection of a sufficiently large number of subjects, simultaneously. As HLA-genotyping is different in the separate studies it was only possible to fulfill the second requirement by downgrading to DR3 and DR4 serotypes used as matching criteria and thus violating condition (1) to some extent. Genotypes LMP2-R/H-DR3/4, LMP2-H/H-DR3/4, LMP7-A/A-DR3/4 and LMP7-A/B-DR3/4 are not specified discretely in Deng et al. [53]. Therefore, we applied the frequency rate to these types as it appeared in the random population to yield discrete values using following formula, considering LMP2-R/H as an example:
F(LMP2-R/H-DR3) = (F(LMP2-R/H-DR3)+F(LMP2-H/H-DR3))·(F(LMP2-R/HR)/(F(LMP2-R/HR)+F(LMP2-H/HR)),
with F(·) as the frequency and R referring to the frequency in the random population. According to this scheme we obtained the following relations: LMP2-R/H-DR3 : LMP2-H/H-DR3 = 0.9 for patients (0.864 for controls) and LMP7-A/A-DR3 : LMP2-A/B-DR3 = 0.401 for patients (0.331 for controls). The same applies for DR4 haplotypes.
Furthermore, HLA-haplotypes are not further characterized to determine homo- and heterozygosity, meaning that at least one chromosome carries the DR3 or DR4 serotype in subjects we include in this analysis. The only study that indicates a specification in this regard is Undlien et al. [69]. The study by Ma and colleagues specifies DR3 and DR4 serotypes in subjects and indicates combinations of DR3/4 positive/negative haplotypes yielding three classifications: 1. DR3+/DR4-, 2. DR3-/DR4- and 3. DR3-/DR4+ [72]. We used the data of the 1. and 3. class of this study to account for DR3 and DR4 positive subjects.
Table 6 finally presents the results from cross-over data with DR3- and DR4-matched individuals. The table contains no data for TAP2*687 as DR-matching spares only data from a single study (van Endert et al. [54]) that are not exactly specified for TAP2*687. Values on TAP2*651 also only includes data from a single study (Penfornis et al. [41]), but these data are specified exactly for TAP and DR. Table 6 contains uncorrected p-values and shows that associations of LMP or TAP genes in the context of HLA linkage are largely insignificant. After correction of p-values only a weak significance remains for LMP2-R/H-DR4 which is twice as frequent in diabetics than in controls (109:51). LMP2-R/R-DR4 and LMP7-B/B-DR4 are significantly less frequent in patients than in healthy controls. Generally, significant differences in the distribution of LMP and TAP genotypes can only be observed in DR4-matched individuals, suggesting that this serotype is more likely a candidate for LD to LMP or TAP.
Table
6.
Standardized residues and significance levels of genotypic associations with HLA-matched T1DM |
|
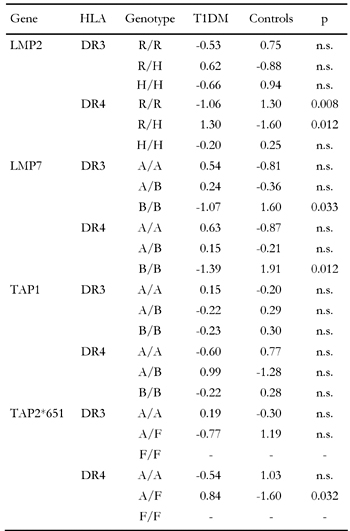 |
 |
Significance levels determined for HLA-matched subjects are lower than in randomly selected (non-HLA-matched) individuals except for LMP2-R/H, which shows approximately the same (weak) degree of association with T1DM. The strong association of TAP2*651-A/F and TAP2*687-A/A with disease is not confirmed in HLA-matched subjects. Taken together, our analysis suggests that the susceptibility conferring genotypes LMP7-A/A, TAP1-A/B, TAP2*651-A/F and TAP2*687-A/A (Table 5) are associated with disease independently of LD. A similar conclusion can be drawn for the protective genotypes. The frequencies of LMP2-R/R, LMP7-B/B, TAP1-A/A, TAP2*651-A/A and TAP2*687-A/B are significantly smaller in randomly selected diabetic patients (Table 5) but only one relation is confirmed in the HLA-matched population, namely LMP2-R/R-DR4 (Table 6), suggesting that protection by allelic variation is conferred largely independently from LD. Analogously to the situation with the susceptibility-conferring genotypes, existence of LD is more likely in association with LMP genes. TAP genes seem to be susceptible or protective in the absence of LD with HLA-DR3 and -DR4.
Conclusion
LMP and TAP genes control the antigen-processing pathway, and polymorphisms in these genes may be responsible for epitope specificity and diversity of self-peptides expressed on APC. Daniel et al. evaluated the peptide affinity of human TAP1 and TAP2 in an artificial neural network and found that HLA class I molecules differed significantly with respect to TAP affinities of their ligands, suggesting that TAP modulates the supply of peptides transported in the ER to associate with newly synthesized MHC class I molecules and thereby contributes to epitope selection [26]. In contrast, the same group showed in an earlier animal experiment that different TAP1 and TAP2 alleles translocated peptides with similar efficiency into the ER which disproves the relevance of polymorphisms in these genes for T1DM development [73]. Discordance also exists regarding the role of the proteasome in its attribution to diabetic pathogenesis [45, 46, 49]. In intracellular antigen processing, proteasome-dependent and -independent pathways are observed making it difficult to interpret which is the preferred mechanism that turns T cells to become autoreactive.
The outcomes of randomized controlled studies that evaluate the genetic susceptibility of LMP and TAP genes for the pathogenesis of T1DM appeared to be unsettled. This has led to the consideration that biases evoked by gene sequence identifying and data collecting methods could be causative. We therefore, pooled data from eleven separate studies and recalculated statistical measurements by the application of genotype and HLA haplotype matching in order to balance such effects. Our results support the consideration that genotypes TAP2*687-A/A and TAP2*651-A/F are associated with diabetes, which has previously been reported by others [40, 50, 64, 65]. We found that this association is independent of LD with DRB1. More surprisingly, it appears that even TAP1-A/B is positively related to disease although this genotype was reported to be only weakly, if at all, associated with disease in other studies [36, 54, 65]. Our analysis also suggests that TAP1-A/A, TAP2*651-A/A and TAP2*687-A/B are the protective types. The acquired association of TAP genes with diabetes appeared not to be secondary to LD with DR3/4. Weak associations with LD were detected in LMP genes. The association of types LMP2-R/H and LMP7-A/A with disease also appeared to be of primary nature. LMP7-B/B was found almost twice as frequently in healthy persons than in patients suggesting that this genotype is protective. In summary, our results suggest that products of particular LMP and TAP allelic variants appear to be involved in rendering antigen-processing pathways to become capable of conferring susceptibility or protection to T1DM. Therefore, if positive selection of autoreactive T cells is critically affected by the diversity of antigenic peptide presentation then we can conclude that TAP and LMP genes have a significant impact on antigen-processing pathways. This, in turn, explains their role in the retention of a balanced immunity. However, it remains to be determined how polymorphisms in these genes providing the diversity of proteins contribute to the formation of epitopes that render CD8+ cells to become autoreactive, and whether these gene products require other factors in conjunction to confer disease.
Acknowledgments:
This work was supported by a grant of the Society for Biomedical Diabetes Research (SBDR).
References
- Townsend A, Ohlen C, Bastin J, Ljunggren HG, Foster L, Karre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature 1989. 340(6233):443-448. [DOD] [CrossRef]
- Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell 1986. 44(6):959-968. [DOD] [CrossRef]
- Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 1991. 351(6324):290-296. [DOD] [CrossRef]
- Kobayashi H, Kimura S, Aoki N, Sato K, Celis E, Katagiri M. Existence of MHC class I-restricted alloreactive CD4+ T cells reacting with peptide transporter-deficient cells. Immunogenetics 2001. 53(8):626-633. [DOD] [CrossRef]
- Boyle LH, Goodall JC, Gaston JS. Major histocompatibility complex class I-restricted alloreactive CD4+ T cells. Immunology 2004. 112(1):54-63. [DOD] [CrossRef]
- Tait BD, Colman PG, Morahan G, Marchinovska L, Dore E, Gellert S, Honeyman MC, Stephen K, Loth A. HLA genes associated with autoimmunity and progression to disease in type 1 diabetes. Tissue Antigens 2003. 61(2):146-153. [DOD]
- Oldstone MB, Nerenberg M, Southern P, Price J, Lewicki H. Virus infection triggers insulin-dependent diabetes mellitus in a transgenic model: role of anti-self (virus) immune response. Cell 1991. 65(2):319-331. [DOD] [CrossRef]
- Kuzushima K, Sun R, van Bleek GM, Vegh Z, Nathenson SG. The role of self peptides in the allogeneic cross-reactivity of CTLs. J Immunol 1995. 155(2):594-601. [DOD]
- Hogquist KA, Gavin MA, Bevan MJ. Positive selection of CD8+ T cells induced by major histocompatibility complex binding peptides in fetal thymic organ culture. J Exp Med 1993. 177(5):1469-1473. [DOD] [CrossRef]
- Imarai M, Goyarts EC, van Bleek GM, Nathenson SG. Diversity of T cell receptors specific for the VSV antigenic peptide (N52-59) bound by the H-2Kb class I molecule. Cell Immunol 1995. 160(1):33-42. [DOD] [CrossRef]
- Boehm BO, Bluestone JA. Differential roles of costimulatory signaling pathways in type 1 diabetes mellitus. Rev Diabetic Stud 2004. 1(4):156-164. [DOD] [CrossRef]
- Huang FP, MacPherson GG. Continuing education of the immune system - dendritic cells, immune regulation and tolerance. Curr Mol Med 2001. 1(4):457-468. [DOD] [CrossRef]
- Ashton-Rickardt PG, Van Kaer L, Schumacher TN, Ploegh HL, Tonegawa S. Peptide contributes to the specificity of positive selection of CD8+ T cells in the thymus. Cell 1993. 73(5):1041-1049. [DOD] [CrossRef]
- Bell JI, Denney D Jr, Foster L, Belt T, Todd JA, McDevitt HO. Allelic variation in the DR subregion of the human major histocompatibility complex. Proc Natl Acad Sci U S A 1987. 84(17):6234-6238. [DOD]
- Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 1987. 329(6140):599-604. [DOD] [CrossRef]
- Ikegami H, Kawaguchi Y, Yamato E, Kuwata S, Tokunaga K, Noma Y, Shima K, Ogihara T. Analysis by the polymerase chain reaction of histocompatibility leucocyte antigen-DR9-linked susceptibility to insulin-dependent diabetes mellitus. J Clin Endocrinol Metab 1992. 75(5):1381-1385. [DOD] [CrossRef]
- Restifo NP, Esquivel F, Asher AL, Stotter H, Barth RJ, Bennink JR, Mule JJ, Yewdell JW, Rosenberg SA. Defective presentation of endogenous antigens by a murine sarcoma. Implications for the failure of an anti-tumor immune response. J Immunol 1991. 147(4):1453-1459. [DOD]
- Joyce S, Mathew JM, Flye MW, Mohanakumar T. A polymorphic human kidney-specific non-MHC alloantigen. Its possible role in tissue-specific allograft immunity. Transplantation 1992. 53(5):1119-1127. [DOD]
- Fedoseyeva EV, Tam RC, Popov IA, Orr PL, Garovoy MR, Benichou G. Induction of T cell responses to a self-antigen following allotransplantation. Transplantation 1996. 61(5):679-683. [DOD] [CrossRef]
- Deverson EV, Gow IR, Coadwell WJ, Monaco JJ, Butcher GW, Howard JC. MHC class II region encoding proteins related to the multidrug resistance family of transmembrane transporters. Nature 1990. 348(6303):738-741. [DOD] [CrossRef]
- van Bleek GM, Nathenson SG. Presentation of antigenic peptides by MHC class I molecules. Trends Cell Biol 1992. 2(7):202-207. [DOD] [CrossRef]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol 1992. 8:67-113. [DOD] [CrossRef]
- Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev 1998. 22(1):1-20. [DOD]
- Elliott T. Transporter associated with antigen processing. Adv Immunol 1997. 65:47-109. [DOD]
- Nijenhuis M, Hammerling GJ. Multiple regions of the transporter associated with antigen processing (TAP) contribute to its peptide binding site. J Immunol 1996. 157(12):5467-5477. [DOD]
- Daniel S, Brusic V, Caillat-Zucman S, Petrovsky N, Harrison L, Riganelli D, Sinigaglia F, Gallazzi F, Hammer J, van Endert PM. Relationship between peptide selectivities of human transporters associated with antigen processing and HLA class I molecules. J Immunol 1998. 161(2):617-624. [DOD]
- Androlewicz MJ, Cresswell P. Human transporters associated with antigen processing possess a promiscuous peptide-binding site. Immunity 1994. 1(1):7-14. [DOD] [CrossRef]
- Anderson K, Cresswell P, Gammon M, Hermes J, Williamson A, Zweerink H. Endogenously synthesized peptide with an endoplasmic reticulum signal sequence sensitizes antigen processing mutant cells to class I-restricted cell-mediated lysis. J Exp Med 1991. 174(2):489-492. [DOD] [CrossRef]
- Bacik I, Cox JH, Anderson R, Yewdell JW, Bennink JR. TAP (transporter associated with antigen processing)-independent presentation of endogenously synthesized peptides is enhanced by endoplasmic reticulum insertion sequences located at the amino- but not carboxyl-terminus of the peptide. J Immunol 1994. 152(2):381-387. [DOD]
- Lewis JW, Elliott T. Evidence for successive peptide binding and quality control stages during MHC class I assembly. Curr Biol 1998. 8(12):717-720. [DOD] [CrossRef]
- Lewis JW, Neisig A, Neefjes J, Elliott T. Point mutations in the alpha 2 domain of HLA-A2.1 define a functionally relevant interaction with TAP. Curr Biol 1996. 6(7):873-883. [DOD] [CrossRef]
- Hearn AR, de Haan L, Pemberton AJ, Hirst TR, Rivett AJ. Trafficking of exogenous peptides into proteasome-dependent major histocompatibility complex class I pathway following enterotoxin B subunit-mediated delivery. J Biol Chem 2004. 279(49):51315-51322. [DOD] [CrossRef]
- Spies T, Bresnahan M, Bahram S, Arnold D, Blanck G, Mellins E, Pious D, DeMars R. A gene in the human major histocompatibility complex class II region controlling the class I antigen presentation pathway. Nature 1990. 348(6303):744-747. [DOD] [CrossRef]
- Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell 1992. 71(7):1205-1214. [DOD] [CrossRef]
- Aldrich CJ, Ljunggren HG, Van Kaer L, Ashton-Rickardt PG, Tonegawa S, Forman J. Positive selection of self- and alloreactive CD8+ T cells in Tap-1 mutant mice. Proc Natl Acad Sci U S A 1994. 91(14):6525-6528. [DOD]
- Faustman D, Li XP, Lin HY, Fu YE, Eisenbarth G, Avruch J, Guo J. Linkage of faulty major histocompatibility complex class I to autoimmune diabetes. Science 1991. 254(5039):1756-1761. [DOD]
- Li F, Guo J, Fu Y, Yan G, Faustman D. Abnormal class I assembly and peptide presentation in the nonobese diabetic mouse. Proc Natl Acad Sci U S A 1994. 91(23):11128-11132. [DOD]
- Kawaguchi Y, Ikegami H, Fukuda M, Takekawa K, Fujioka Y, Fujisawa T, Ueda H, Ogihara T. Absence of association of TAP and LMP genes with type 1 (insulin-dependent) diabetes mellitus. Life Sci 1994. 54(26):2049-2053. [DOD] [CrossRef]
- Chauffert M, Cisse A, Chevenne D, You JF, Michel S, Trivin F. Susceptibility to type 1 diabetes in the Senegalese population is linked to HLA-DQ and not TAP and LMP genes. Diabetes Care 1997. 20(8):1299-1303. [DOD]
- Jackson DG, Capra JD. TAP2 association with insulin-dependent diabetes mellitus is secondary to HLA-DQB1. Hum Immunol 1995. 43(1):57-65. [DOD] [CrossRef]
- Penfornis A, Tuomilehto-Wolf E, Faustman DL, Hitman GA. Analysis of TAP2 polymorphisms in Finnish individuals with type I diabetes. DiMe (Childhood Diabetes in Finland) Study Group. Hum Immunol 2002. 63(1):61-70. [DOD] [CrossRef]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995. 268(5210):533-539. [DOD]
- Groll M, Coux O. Proteasomes in proteinase and peptidase inhibition: recent potential targets for drug development. In: Smith HJ, Simons C (eds.), Taylor & Francis Publishers, NY, USA, 2002, pp. 62-84. [DOD]
- Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 2005. 57(1-2):33-41. [DOD] [CrossRef]
- Gaczynska M, Rock KL, Goldberg AL. Role of proteasomes in antigen presentation. Enzyme Protein 1993. 47(4-6):354-369. [DOD]
- Michalek MT, Grant EP, Gramm C, Goldberg AL, Rock KL. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature 1993. 363(6429):552-554. [DOD] [CrossRef]
- Sibille C, Gould KG, Willard-Gallo K, Thomson S, Rivett AJ, Powis S, Butcher GW, De Baetselier P. LMP2+ proteasomes are required for the presentation of specific antigens to cytotoxic T lymphocytes. Curr Biol 1995. 5(8):923-930. [DOD] [CrossRef]
- Fehling HJ, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, von Boehmer H. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science 1994. 265(5176):1234-1237. [DOD]
- van Kaer L, Ashton-Rickardt PG, Eichelberger M, Gaczynska M, Nagashima K, Rock KL, Goldberg AL, Doherty PC, Tonegawa S. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity 1994. 1(7):533-541. [DOD] [CrossRef]
- Zhou P, Cao H, Smart M, David C. Molecular basis of genetic polymorphism in major histocompatibility complex-linked proteasome gene (Lmp-2). Proc Natl Acad Sci U S A 1993. 90(7):2681-2684. [DOD]
- Momburg F, Ortiz-Navarrete V, Neefjes J, Goulmy E, van de Wal Y, Spits H, Powis SJ, Butcher GW, Howard JC, Walden P, et al. Proteasome subunits encoded by the major histocompatibility complex are not essential for antigen presentation. Nature 1992. 360(6400):174-177. [DOD] [CrossRef]
- Yewdell J, Lapham C, Bacik I, Spies T, Bennink J. MHC-encoded proteasome subunits LMP2 and LMP7 are not required for efficient antigen presentation. J Immunol 1994. 152(3):1163-1170. [DOD]
- Deng GY, Muir A, Maclaren NK, She JX. Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: population and family studies. Am J Hum Genet 1995. 56(2):528-534. [DOD]
- van Endert PM, Liblau RS, Patel SD, Fugger L, Lopez T, Pociot F, Nerup J, McDevitt HO. Major histocompatibility complex-encoded antigen processing gene polymorphism in IDDM. Diabetes 1994. 43(1):110-117. [DOD]
- Dyall R, Messaoudi I, Janetzki S, Nikolic-Zugic J. MHC polymorphism can enrich the T cell repertoire of the species by shifts in intrathymic selection. J Immunol 2000. 164(4):1695-1698. [DOD]
- Ashton-Rickardt PG, Bandeira A, Delaney JR, Van Kaer L, Pircher HP, Zinkernagel RM, Tonegawa S. Evidence for a differential avidity model of T cell selection in the thymus. Cell 1994. 76(4):651-663. [DOD] [CrossRef]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell 1994. 76(1):17-27. [DOD] [CrossRef]
- Mathews CE, Pietropaolo SL, Pietropaolo M. Reduced thymic expression of islet antigen contributes to loss of self-tolerance. Ann N Y Acad Sci 2003. 1005:412-417. [DOD] [CrossRef]
- Nikolich-Zugich J, Fremont DH, Miley MJ, Messaoudi I. The role of mhc polymorphism in anti-microbial resistance. Microbes Infect 2004. 6(5):501-512. [DOD] [CrossRef]
- Chen M, Abele R, Tampe R. Peptides induce ATP hydrolysis at both subunits of the transporter associated with antigen processing. J Biol Chem 2003. 278(32):29686-29692. [DOD] [CrossRef]
- van Bleek GM, Nathenson SG. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature 1990. 348(6298):213-216. [DOD] [CrossRef]
- van Bleek GM, Nathenson SG. The structure of the antigen-binding groove of major histocompatibility complex class I molecules determines specific selection of self-peptides. Proc Natl Acad Sci U S A 1991. 88(24):11032-11036. [DOD]
- Grossman Z, Min B, Meier-Schellersheim M, Paul WE. Concomitant regulation of T-cell activation and homeostasis. Nat Rev Immunol 2004. 4(5):387-395. [DOD] [CrossRef]
- Marsh SG, Bodmer JG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Hansen JA, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI. Nomenclature for factors of the HLA system, 2000. Tissue Antigens 2001. 57(3):236-283. [DOD] [CrossRef]
- Colonna M, Bresnahan M, Bahram S, Strominger JL, Spies T. Allelic variants of the human putative peptide transporter involved in antigen processing. Proc Natl Acad Sci U S A 1992. 89(9):3932-3936. [DOD]
- Caillat-Zucman S, Bertin E, Timsit J, Boitard C, Assan R, Bach JF. Protection from insulin-dependent diabetes mellitus is linked to a peptide transporter gene. Eur J Immunol 1993. 23(8):1784-1788. [DOD]
- Jackson DG, Capra JD. TAP1 alleles in insulin-dependent diabetes mellitus: a newly defined centromeric boundary of disease susceptibility. Proc Natl Acad Sci U S A 1993. 90(23):11079-11083. [DOD]
- Ronningen KS, Undlien DE, Ploski R, Maouni N, Konrad RJ, Jensen E, Hornes E, Reijonen H, Colonna M, Monos DS, et al. Linkage disequilibrium between TAP2 variants and HLA class II alleles; no primary association between TAP2 variants and insulin-dependent diabetes mellitus. Eur J Immunol 1993. 23(5):1050-1056. [DOD]
- Undlien DE, Akselsen HE, Joner G, Dahl-Jorgensen K, Sovik O, Ronningen KS, Thorsby E. No independent associations of LMP2 and LMP7 polymorphisms with susceptibility to develop IDDM. Diabetes 1997. 46(2):307-312. [DOD]
- Ding H, Cheng H, Fu Z, Yan L, Yang G. Relationship of large multifunctional proteasome 7 gene polymorphism with susceptibility to type 1 diabetes mellitus and DR3 gene. Chin Med J (Engl) 2001. 114(12):1263-1266. [DOD]
- Ding HL, Cheng H, Fu ZZ, Deng QL, Tang Yan L, Yan T. The relationship of lmp2 and DR3 genes with susceptibility to type I diabetes mellitus in south China Han population. World J Gastroenterol 2000. 6(1):111-114. [DOD]
- Ma L, Penfornis A, Wang X, Schoenfeld D, Tuomilehto-Wolf E, Metcalfe K, Hitman G, Faustman D. Evaluation of TAP1 polymorphisms with insulin dependent diabetes mellitus in Finnish diabetic patients. The Childhood Diabetes in Finland (DiMe) Study Group. Hum Immunol 1997. 53(2):159-166. [DOD] [CrossRef]
- Daniel S, Caillat-Zucman S, Hammer J, Bach JF, van Endert PM. Absence of functional relevance of human transporter associated with antigen processing polymorphism for peptide selection. J Immunol 1997. 159(5):2350-2357. [DOD]
This article has been cited by other articles:
|
The type 1 diabetes - HLA susceptibility interactome - identification of HLA genotype-specific disease genes for type 1 diabetes
Brorsson C, Tue Hansen N, Bergholdt R, Brunak S, Pociot F
PLoS One 2010. 5(3):e9576
|
|

|
Long-term and acute effects of gliadin on small intestine of patients on potentially pathogenic networks in celiac disease
Castellanos-Rubio A, Santin I, Martin-Pagola A, Irastorza I, Castano L, Vitoria JC, Bilbao JR
Autoimmunity 2010. 43(2):131-139
|
|

|
Genetic deficiency of Itgb2 or ItgaL prevents autoimmune diabetes through distinctly different mechanisms in NOD/LtJ mice
Glawe JD, Patrick DR, Huang M, Sharp CD, Barlow SC, Kevil CG
Diabetes 2009. 58(6):1292-1301
|
|

|
HLA and infectious diseases
Blackwell JM, Jamieson SE, Burgner D
Clin Microbiol Rev 2009. 22(2):370-385
|
|

|
Multiple Drug Resistance Associated with Function of ABC-Transporters in Diabetes Mellitus: Molecular Mechanism and Clinical Relevance
Koehn J, Fountoulakis M, Krapfenbauer K
Infect Disord Drug Targets 2008. 8(2):109-118
|
|
|
Prediction of autoimmune diseases: myth, reality and risk
Ortega-Hernandez OD, Lopez-Guzman S, Rojas-Villarraga A, Anaya JM
Rev Fac Med 2008. 16(1):56-73
|
|

|
Death Pathways in T Cell Homeostasis and Their Role in Autoimmune Diabetes
Gronski MA, Weinem M
Rev Diabet Stud 2006. 3(2):88-95
|
|

|
The role of HLA class I gene variation in autoimmune diabetes
Sia C, Weinem M
Rev Diabet Stud 2005. 2(2):97-109
|
|
|