Original Data
| Rev Diabet Stud,
2008,
5(1):28-37 |
DOI 10.1900/RDS.2008.5.28 |
Adiponectin and Adiponectin Receptor Gene Variants in Relation to Type 2 Diabetes and Insulin Resistance-Related Phenotypes
Viktor A. Potapov1, Dimitry A. Chistiakov1, Anna Dubinina2, Minara S. Shamkhalova2, Marina V. Shestakova2, Valery V. Nosikov1
1National Research Center GosNIIgenetika, 117545 Moscow, Russia
2Endocrinological Research Center, 117036 Moscow, Russia
Address correspondence to: Dimitry A. Chistiakov, e-mail: dimitry.chistiakov@eudoramail.com
Manuscript submitted April 15, 2008; resubmitted May 22, 2008; accepted May 25, 2008.
Keywords: type 2 diabetes, adiponectin, insulin resistance, haplotype, ADIPOQ, ADIPOR1, ADIPOR2
Abstract
BACKGROUND: Alterations in adiponectin-mediated pathways are known to be associated with glucose intolerance, insulin resistance (IR), obesity, and type 2 diabetes (T2D) mellitus. Genetic variations in adiponectin (ADIPOQ) and adiponectin 1 and 2 receptor (ADIPOR1 and ADIPOR2) could have effects on IR-related phenotypes and T2D. Here we examine whether the polymorphic markers rs2241766 (ADIPOQ), rs22753738 (ADIPOR1), rs11061971 and rs16928751 (both in ADIPOR2) are implicated in susceptibility to T2D in a Russian population. METHODS: The polymorphic markers were genotyped in 129 T2D patients, and 117 non-diabetic controls, by polymerase chain reaction (PCR) restriction fragment length polymorphism approach. In the subjects, biochemical characteristics including serum insulin, plasma glucose and serum lipids/lipoproteins were measured and compared for correlation with the genetic variations studied. RESULTS: Allele T of rs11061971 and allele A of rs16928751 showed association with higher risk of diabetes providing odds ratios (OR) of 2.05 (p = 0.0025) and 1.88 (p = 0.018), respectively. Haplotype A-G consisting of allele A of rs11061971 and allele G of rs16928751 was associated with reduced risk of T2D (OR = 0.59, pc = 0.0224). Compared to other variants, diabetic patients double homozygous for A/A of rs16928751 and G/G of rs16928751 had decreased homeostasis model assessment-insulin resistance (pc = 0.0375) and serum triglycerides (pc = 0.0285). CONCLUSIONS: The variants of ADIPOR2 confer susceptibility to T2D and are associated with some IR-related phenotypes in the Russian study population.
Introduction
Adiponectin, a product of the ADIPOQ (APM1) gene, is the most abundant human adipose- specific protein [1]. Decreased levels of plasma adiponectin and adiponectin mRNA were observed in both patients with T2D and ob/ob mice, an obese diabetic murine model, suggesting downregulation of the adiponectin expression in adipocytes in obesity and T2D [2-4]. Reduced adiponectin is associated with decreased lipid oxidation, increased triglyceride content and suppression of insulin-dependent signaling in peripheral tissues such as liver and muscle. By this process decreased adiponectin can contribute to insulin resistance (IR) and obesity [5].
The ADIPOQ gene comprising 3 exons spans 16 kb on chromosome 3q27 [4]. Correlations of adiponectin levels and phenotypes associated with the metabolic syndrome and T2D, such as insulin resistance, glucose intolerance and high density lipoprotein/cholesterol ratio, have been reported for several polymorphic markers including a thymine-to-guanine substitution at position +45 (+45 T/G; rs2241766) in exon 2 of ADIPOQ [5]. Multiple associations of this kind observed for the ADIPOQ +45 T/G variant suggest that in principle this single nucleotide polymorphism (SNP) represents a useful marker to evaluate the relationship between the ADIPOQ gene, T2D and quantitative characteristics of the metabolic syndrome.
Two receptor forms, AdipoR1 and AdipoR2, mediate biologic effects of adiponectin. In humans, AdipoR1 is ubiquitously expressed, with highest levels being in the skeletal muscle. AdipoR2 is predominantly expressed in skeletal muscle and the liver [6]. Expression of adiponectin receptors was also found in pancreatic β-cells, and fatty acids may regulate their expression levels in β-cells [7]. In mice, simultaneous disruption of both AdipoR1 and R2 abolished adiponectin binding with consequential actions resulting in greater tissue triglyceride content, inflammation, and oxidative stress, thereby leading to insulin resistance and marked glucose intolerance [8]. The expression of both receptors was shown to be substantially reduced in the skeletal muscle of T2D patients and insulin-resistant T2D obese db/db mice [9, 10]. Indeed, low levels of AdipoR in peripheral tissues in obesity and T2D might reduce biological effects of adiponectin and therefore further aggravate the negative metabolic effect of low levels of adiponectin which characterizes both obese and diabetic states.
Since AdipoR1 and AdipoR2 mediate the effects of adiponectin on target tissues, they have been considered to be strong candidates in the risk of type 2 diabetes [11]. The ADIPOR1 gene located on chromosome 1p36.13-q41 contains 8 exons [12]. The (-106) T/G SNP (rs22753738) is situated in intron 1. In Finnish glucose intolerant patients, this marker showed an association with size characteristics (weight, waist circumference, saggital diameter) [13], i.e. the indicators of central obesity (body size), which are frequently linked to insulin resistance and metabolic syndrome irrespective of height or BMI [14].
Similarly to ADIPOR1, ADIPOR2, which resides on chromosome 12p13.31, also contains 8 exons [12]. Some ADIPOR2 variants showed association with insulin resistance-related phenotypes such as decreased fasting triglyceride levels [15] and higher respiratory quotient, i.e. lower rate of fat oxidation [16]. At present, the reported results for association between ADIPOR2 and T2D for several Caucasian populations show some inconsistency [11, 17-19]. Due to the controversy in association between ADIPOR2 and T2D, we intended to evaluate whether this gene is also associated with T2D in a Russian population. For analysis, we selected two common polymorphisms: rs11061971 (+219 A/T) and rs16928751 (+795 G/A) located respectively in intron 2 and exon 6 of ADIPOR2. The marker rs11061971 showed association with both glucose intolerance (p = 0.021) and T2D (p = 0.001) in Old Order Amish [17]. In the present study, we also evaluated two more markers, rs22753738 and rs2241766 SNPs, located in ADIPOR1 and ADIPOQ respectively, for association with T2D.
Subjects and Methods
Subjects
A total of 129 Russian unrelated patients with T2D and 117 Russian non-diabetic controls were studied. The subjects were recruited in the Endocrinology Research Center. The T2D patients were selected according to the World Health Organization diagnostic criteria as having either, a fasting plasma glucose concentration ≥ 7.8 mmol/l, or a plasma glucose concentration ≥ 11.1 mmol/l 2 h after a 75-g oral glucose intake [20]. All study participants resided in Moscow or the Moscow region. The controls were normoglycemic; no past history of glucose intolerance; an HbA1c level of < 6.4%, or a normal oral glucose tolerance test (OGTT); and no family history of diabetes. To avoid interferences of biological variables, controls with previous diagnosis of type 1 diabetes, or those receiving treatment for hypercholesterolemia, hypertension or T2D, were excluded from the study. The study protocol was approved by the Review Board of the Endocrinology Research Center in Moscow, Russia, and all participants gave written informed consent.
Biochemical measurements
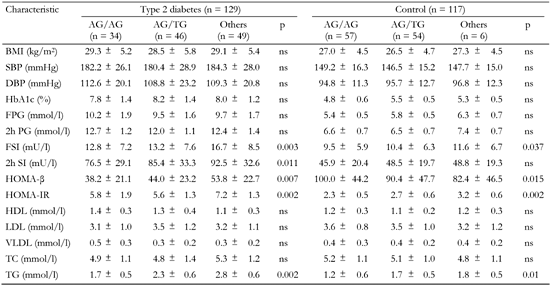
Blood was drawn for fasting cholesterol, high-density lipoprotein (HDL), cholesterol, triglycerides, plasma glucose and insulin after 12-h overnight fast. These parameters were measured by standard enzymatic assays. Low-density lipoprotein (LDL) cholesterol was derived using the Friedewald equation [21]. A standard 75-g OGTT was performed after a 12-h overnight fast according to the WHO recommendations. HbA1c was measured using ion-exchange high performance liquid chromatography (normal reference range: 4.1-6.4%). Plasma insulin levels were determined by means of an enzymatic immunoassay. Homeostasis model assessment of β-cell function (HOMA-β) was calculated as 20 × fasting plasma insulin (mU/l) divided by (fasting glucose (mmol/l) - 3.5). Homeostasis model assessment-insulin resistance (HOMA-IR) was computed as fasting plasma insulin (mU/l) × fasting glucose (mmol/l)/ 22.5 [22]. Clinical and metabolic characteristics of diabetic patients and non-affected individuals are summarized in Table 1.
DNA analysis
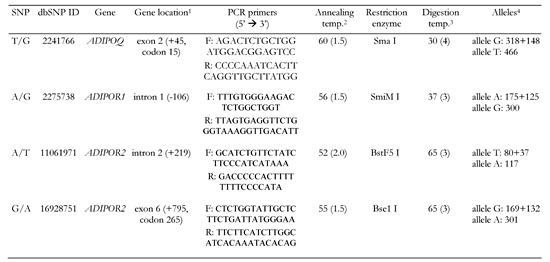
Total DNA was isolated from whole-blood samples pretreated with proteinase K, using a standard protocol for extraction with phenol-chloroform (Fermentas, Vilnius, Lithuania). For each gene studied, polymorphic regions were amplified by a polymerase chain reaction (PCR) in a total volume of 25 μl, using PCR reagents and Taq polymerase from Fermentas followed by digestion of a PCR product with a corresponding restriction enzyme. PCR- restriction fragment length polymorphism (RFLP) assays for detecting each SNP are described in Table 2. Restriction enzymes used in these assays were manufactured in Sybenzyme (Novosibirsk, Russia). PCR buffer consisted of 10 mmol/l Tris-HCl, pH 8.8, 50 mmol/l KCl, 0.08% Nonidet P40, 1.5 mmol/l MgCl2, 0.2 mmol/l of each dNTPs, 100 ng of genomic DNA, 1 U Taq polymerase and 10 pmoles of each primer. PCR cycling was performed in a DNA Engine Dyad Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) as follows: initial denaturation step at 96°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing for 30 s and extension at 72°C for 30 s, with a final extension of 7 min at 72°C. 5 μl of the PCR product was then cleaved with a corresponding restriction enzyme.
Restriction products were analyzed by electrophoresis either in 2% agarose gel (for rs2241766, rs2275738, and rs16928751 SNPs) or in 8% polyacrylamide gel (for rs11061971). To visualize the digestion fragments, an agarose gel was then stained with ethidium bromide and exposed under UV, whereas a polyacrylamide gel was stained with silver nitrate.
Statistical analysis
Data were analyzed with the SPSS/Win programs (version 10.0; SPSS Inc., Chicago, IL, USA). Results are given as mean ± SD or percentages. The clinical and laboratory characteristics of the T2D patients and controls were compared with the unpaired Student's t-test or χ2 test, as appropriate. To compare quantitative data in three groups, a one-way ANOVA test was used. The degree of pairwise linkage disequilibrium (LD) between markers was calculated using LDA software [23]. The test for Hardy-Weinberg equilibrium and comparison of genotype and allele frequencies in the T2D subjects and controls were performed using the χ2 test. Odds ratios (OR) and their 95% confidence intervals (CI) were calculated, using the Calculator for Confidence Intervals of Odds Ratio [24]. ORs were computed when data were significantly different. After correction for multiple comparisons, p-values (pc) of less then 0.05 were considered significant.
Results
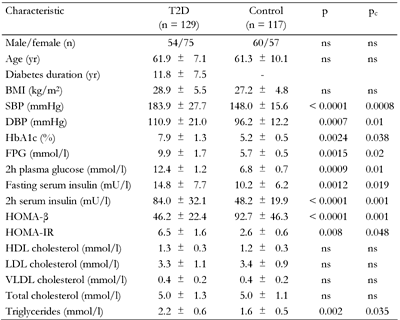
Compared to the controls, diabetic patients had significantly elevated blood pressure, glycosylated hemoglobin HbA1c, triglyceride and both (fasting and 2h) insulin and glucose levels in blood (Table 1). These data could reflect presence of hypertension, hyperglycemia, hyperglyceridemia, altered insulin response and glucose intolerance, i.e. typical T2D-related phenotypes in affected patients. In affected subjects, HOMA-β was significantly lower then in non-diabetic individuals (46.2 ± 22.4 vs. 92.7 ± 46.3, pc < 0.005), indicating β-cell dysfunction in these patients. In contrast, HOMA-IR was significantly increased in T2D patients (6.5 ± 1.6 vs. 2.6 ± 0.6, pc < 0.05) pointing to insulin resistance (Table 1).
Table
1.
Clinical and metabolic characteristics of type 2 diabetes patients and control subjects |
|
 |
 |
Legend:
Data are mean ± SD calculated by chi-squared test. T2D: type 2 diabetes. BMI: body mass index. SBP: systolic blood pressure. DBP: diastolic blood pressure. FPG: fasting plasma glucose. HOMA-β: homeostasis model assessment of β-cells. HOMA-IR: homeostasis model assessment of insulin resistance. HDL: high-density lipoprotein cholesterol. LDL: low-density lipoprotein cholesterol. VLDL: very low-density lipoprotein cholesterol. ns: not significant. |
|
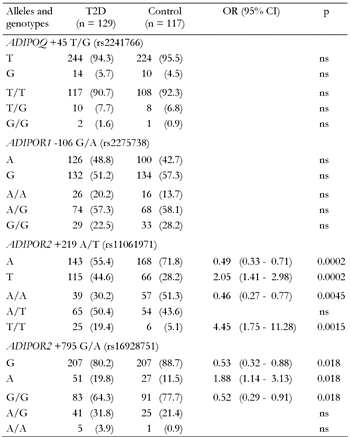
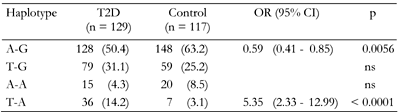
All polymorphic markers studied followed the Hardy-Weinberg equilibrium (data not shown). For the ADIPOQ +45 T/G and ADIPOR1 -106 G/A variants, there were no significant differences in allele frequencies between diabetic and non-diabetic patients (Table 3). However, both markers within the ADIPOR2 gene showed association with T2D. Allele +219T of rs11061971 and allele +795A of rs16928751 were significantly associated with higher risk of diabetes (OR of 2.05 and 1.88, respectively). Both ADIPOR2 markers are in strong LD (D' = 0.79, p < 0.05). The SNP was found to share a common risk haplotype that consisted of the +219T and +795A alleles (OR = 5.35, p < 0.001; Table 4). In contrast, the haplotype of ADIPOR2 comprising the +219A and +795G alleles was associated with lower T2D risk (OR = 0.59, p < 0.01).
Table
2.
Detection of single nucleotide polymorphisms (SNPs) within the ADIPOQ, ADIPOR1 and ADIPOR2 genes |
|
 |
 |
Legend:
1 Position of the translation start (bp). 2 Temperature in °C, Mg2+ in mM in parentheses. 3 Temperature in °C, duration of diagestion in h in parentheses. 4 Definition of alleles and length of digestion products in bp. |
|
Table
3.
Association of the ADIPOQ, ADIPOR1 and ADIPOR2 gene variants with T2D |
|
 |
 |
Legend:
Frequencies are absolute numbers (percentages in parentheses). ns: not significant. |
|
Table
4.
Association of ADIPOR2 haplotypes with T2D |
|
 |
 |
Legend:
Frequencies are absolute numbers (percentages in parentheses). ns: not significant. |
|
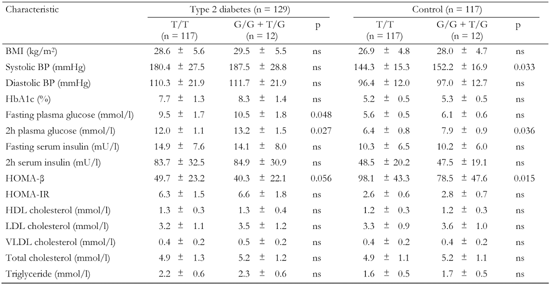
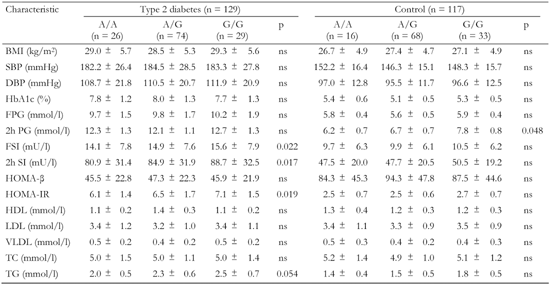
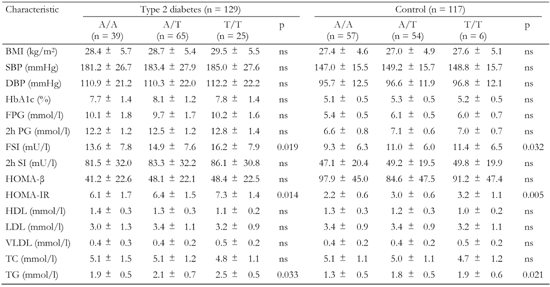
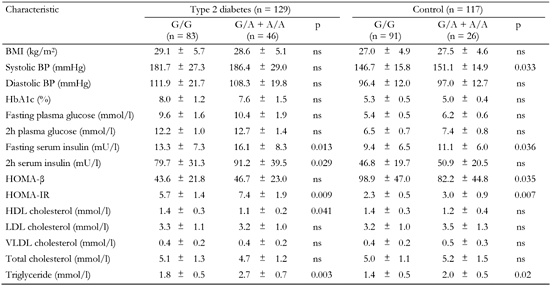
The polymorphic markers showed no association with metabolic characteristics except for the ADIPOR2 +795 G/A variant, which was associated with serum triglyceride levels (Tables A1-A4, see Appendix). Diabetic patients homozygous for G/G had significantly lower triglycerides compared to G/A+A/A (1.8 mmol/l vs. 2.7 mmol/l, pc < 0.05; Table A4, see Appendix). We compared metabolic characteristics between carriers of common ADIPOR2 haplogenotypes occurring at a frequency of more than 10%, to evaluate whether ADIPOR2 haplotypes that showed association with T2D are also associated with IR-related phenotypes. In the serum of T2D patients who were double homozygous for +219 A/A and +795 G/G of ADIPOR2, concentrations of triglycerides were found to be significantly reduced compared with other variants (Table A5). In both diabetic and non-diabetic individuals double homozygotes for +219 A/A and +795 G/G of ADIPOR2, HOMA-IR values were also significantly lower than those in carriers of other diplotypes. These findings suggest a protective role for the haplotype AG of ADIPOR2 in the development of IR.
Discussion
In this study, we examined whether sequence variations in the ADIPOQ and ADIPOR genes contributed to susceptibility to T2D and variations in T2D-related metabolic characteristics in a Russian population. Both markers of ADIPOR2, rs11061971 (+219 A/T) and rs16928751 (+795 G/A), showed association with T2D (Table 3). Similarly, Dumcott et al. reported that the allele T of rs11061971 was significantly associated with higher risk of T2D (OR = 1.47 and 1.71 for T2D vs. normoglycemic patients and T2D + glucose-intolerant vs. normoglycemic patients, respectively) [25]. This SNP is in high LD with several disease-associated polymorphisms that reside in the entire LD block spanning nearly 100 kb across a whole ADIPOR2 gene from the promoter region until the beginning of exon 8 [25, 26].
In the Russian population, the +219 A/T marker was shown to be in strong LD with the +795 G/A SNP of ADIPOR2. The alleles +219A and +795 G forming the common haplotype AG were associated with reduced risk of T2D. The protective role of this haplotype in the development of T2D could be attributed to its association with a decreased HOMA-IR value and therefore with reduced IR. In addition, the haplotype AG showed an association with lower serum concentrations of triglycerides. Other studies also found the relationship between multiple ADIPOR2 variants and triglyceride levels [27, 28]. Tirosh et al. suggested that increased serum triglycerides (>1.7 mmol/l) in combination with elevated fasting glucose could substantially increase the risk for early development of T2D [29].
Compared to ADIPOR1, an emerging role for ADIPOR2 in T2D, as observed in our study, could arise from the unique function of this receptor in mediating effects of adiponectin in the liver. This organ was suggested as the primary site of adiponectin bioactivity [30]. Adiponectin influences fat metabolism increasing fatty acid oxidation through activation of AMP-activated protein kinase, which in turn phosphorylates acetyl CoA carboxylase [31]. Adiponectin also decreases hepatic glucose output by reducing the expression of enzymes involved in gluconeogenesis in the liver [32]. Therefore, increased serum triglycerides could reflect a low lipid oxidation rate in patients with metabolic syndrome, leading to the accumulation of toxic intracellular lipid metabolites in the liver and other peripheral tissues, and hence inhibit insulin signaling [33].
Recently, the development of AdipoR2-/- mice was reported [34]. Disruption of AdipoR2 abolished pancreatic β-cell replication and neogenesis specifically stimulated by insulin resistance. As a consequence, at high-fat feeding, AdipoR2-deficient mice developed persistent hyperglycemia and overt diabetes [34]. Disruption of AdipoR2 in mice very closely mimics the development of human T2D with two key pathophysiological features: IR and inability of pancreatic β-cells to compensate. This evidence highlights a role of AdipoR2 in the pathogenesis of IR syndrome and T2D. Genetic associations observed between certain variants of ADIPOR2 and T2D in several studies, including this one, suggest AdipoR2 as a promising target for the treatment of T2D patients, particularly those who have adiposity, insulin resistance and dyslipidemia.
Appendix
Table
A1.
Association of the ADIPOQ +45 T/G gene variant with metabolic characteristics in T2D patients and non-diabetic controls |
|
 |
 |
Legend:
Data are mean ± SD. BMI: body mass index. BP: blood pressure. HOMA-β: homeostasis model assessment of β-cells. HOMA-IR: homeostasis model assessment of insulin resistance. HDL: high-density lipoprotein cholesterol. LDL: low-density lipoprotein cholesterol. VLDL: very low-density lipoprotein cholesterol. ns: not significant. |
|
Table
A2.
Association of the ADIPOR1 -106 G/A gene variant with metabolic characteristics in T2D patients and non-diabetic controls |
|
 |
 |
Legend:
Data are mean ± SD. BMI: body mass index. SBP: systolic blood pressure. DBP: diastolic blood pressure. FPG: fasting plasma glucose. FSI: fasting serum insulin. HOMA-β: homeostasis model assessment of β-cells. HOMA-IR: homeostasis model assessment of insulin resistance. HDL: high-density lipoprotein cholesterol. LDL: low-density lipoprotein cholesterol. VLDL: very low-density lipoprotein cholesterol. TC: total cholesterol. TG: triglyceride. |
|
Table
A3.
Association of the +219 A/T polymorphism of ADIPOR2 with metabolic characteristics in T2D patients and non-diabetic controls |
|
 |
 |
Legend:
Data are mean ± SD. BMI: body mass index. SBP: systolic blood pressure. DBP: diastolic blood pressure. FPG: fasting plasma glucose. FSI: fasting serum insulin. HOMA-β: homeostasis model assessment of β-cells. HOMA-IR: homeostasis model assessment of insulin resistance. HDL: high-density lipoprotein cholesterol. LDL: low-density lipoprotein cholesterol. VLDL: very low-density lipoprotein cholesterol. TC: total cholesterol. TG: triglyceride. |
|
Table
A4.
Association of the +795 G/A polymorphism of ADIPOR2 with metabolic characteristics in T2D patients and non-diabetic controls |
|
 |
 |
Legend:
Data are mean ± SD. BMI: body mass index. BP: blood pressure. HOMA-β: homeostasis model assessment of β-cells. HOMA-IR: homeostasis model assessment of insulin resistance. HDL: high-density lipoprotein cholesterol. LDL: low-density lipoprotein cholesterol. VLDL: very low-density lipoprotein cholesterol. |
|
Table
A5.
Association of ADIPOR2 haplotype combinations with metabolic characteristics in T2D patients and non-diabetic controls |
|
 |
 |
Legend:
Data are mean ± SD. BMI: body mass index. SBP: systolic blood pressure. DBP: diastolic blood pressure. FPG: fasting plasma glucose. FSI: fasting serum insulin. HOMA-β: homeostasis model assessment of β-cells. HOMA-IR: homeostasis model assessment of insulin resistance. HDL: high-density lipoprotein cholesterol. LDL: low-density lipoprotein cholesterol. VLDL: very low-density lipoprotein cholesterol. TC: total cholesterol. TG: triglyceride. |
|
References
- Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (adipose most abundant gene transcript 1). Biochem Biophys Res Commun 1996. 221:286-289. [DOD] [CrossRef]
- Hotta K, Funahashi T, Arita Y, Takahashi M, Matsuda M, Okamoto Y, Iwanashi H, Kuriyama H, Ouchi N, Maeda K, et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol 2000. 20:1595-1599. [DOD]
- Statnick MA, Beavers LS, Conner LJ, Corominola H, Johnson D, Hammond CD, Rafaeloff-Phail R, Seng T, Suter TM, et al. Decreased expression of apM1 in omental and subcutaneous adipose tissue of humans with type 2 diabetes. Int J Exp Diabetes Res 2000. 1:81-88. [DOD] [CrossRef]
- Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose specific gene dysregulated in obesity. J Biol Chem 1996. 271:10697-10703. [DOD] [CrossRef]
- Vasseur F, Meyre D, Froguel P. Adiponectin, type 2 diabetes and the metabolic syndrome: lessons from human genetic studies. Expert Rev Mol Med 2006. 8:1-12. [DOD]
- Kadowaki T, Yamauchi T. Adiponectin and adiponectin receptors. Endocr Rev 2005. 26:439-451. [DOD] [CrossRef]
- Kharroubi I, Rasschaert J, Eizirik DL, Cnop M. Expression of adiponectin receptors in pancreatic cells. Biochem Biophys Res Commun 2003. 312:1118-1122. [DOD] [CrossRef]
- Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, Iwabu M, Okada-Iwabu M, Kawamoto S, Kubota N, Kubota T, et al. Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 2007. 13:332-339. [DOD] [CrossRef]
- Debard C, Laville M, Berbe V, Loizon E, Guillet C, Morio-Liondore B, Boirie Y, Vidal H. Expression of key genes of fatty acid oxidation, including adiponectin receptors, in skeletal muscle of type 2 diabetic patients. Diabetologia 2004. 47:917-925. [DOD] [CrossRef]
- Inukai K, Nakashima Y, Watanabe M, Takata N, Sawa T, Kurihara S, Awata T, Katayama S. Regulation of adiponectin receptor gene expression in diabetic mice. Am J Physiol Endocrinol Metab 2005. 288:E876-E882. [DOD] [CrossRef]
- Vaxillaire M, Dechaume A, Vasseur-Delannoy V, Lahmidi S, Vatin V, Lepretre F, Boutin P, Hercberg S, Charpentier G, Dina C, et al. Genetic analysis of ADIPOR1 and ADIPOR2 candidate polymorphisms for type 2 diabetes in the Caucasian population. Diabetes 2006. 55:856-861. [DOD] [CrossRef]
- Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 2003. 423:762-769. [DOD] [CrossRef]
- Siitonen N, Pulkkinen L, Mager U, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaaniemi S, Tuomilehto J, et al. Association of sequence variations in the gene encoding adiponectin receptor 1 (ADIPOR1) with body size and insulin levels. The Finnish Diabetes Prevention Study. Diabetologia 2006. 49:1795-1805. [DOD] [CrossRef]
- Kahn R, Buse J, Ferrannini E, Stern M. The metabolic syndrome: time for a critical appraisal. Joint statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 2005. 48:1684-1699. [DOD] [CrossRef]
- Richardson DK, Schneider J, Fourcaudot MJ, Rodriguez LM, Arya R, Dyer TD, Almasy L, Blangero J, Stern MP, DeFronzo RA, et al. Association between variants in the genes for adiponectin and its receptors with insulin resistance syndrome (IRS)-related phenotypes in Mexican Americans. Diabetologia 2006. 49:2317-2328. [DOD] [CrossRef]
- Loos RJ, Ruchat S, Rankinen T, Tremblay A, Perusse L, Bouchard C. Adiponectin and adiponectin receptor gene variants in relation to resting metabolic rate, respiratory quotient, and adiposity-related phenotypes in the Quebec Family Study. Am J Clin Nutr 2007. 85:26-34. [DOD]
- Damcott CM, Ott SH, Pollin TI, Reihart LJ, Wang J, O'Connell JR, Mitchell BD, Shuldiner AR. Genetic variation in adiponectin receptor 1 and adiponectin receptor 2 is associated with type 2 diabetes in the Old Order Amish. Diabetes 2005. 54:2245-2250. [DOD] [CrossRef]
- Ruchat SM, Loos RJ, Rankinen T, Vohl MC, Weisnagel SJ, Despres JP, Bouchard C, Perusse L. Associations between glucose tolerance, insulin sensitivity and insulin secretion phenotypes and polymorphisms in adiponectin and adiponectin receptor genes in the Quebec Family Study. Diabet Med 2008. 25:400-406. [DOD] [CrossRef]
- Qi L, Doria A, Giorgi E, Hu FB. Variations in adiponectin receptor genes and susceptibility to type 2 diabetes in women: a tagging-single nucleotide polymorphism haplotype analysis. Diabetes 2007. 56:1586-1591. [DOD] [CrossRef]
- WHO. Diabetes mellitus. Report of a WHO Study Group. World Health Organ Tech Rep Ser 1985. 727:1-113. [DOD]
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972. 18:499-502. [DOD]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985. 28:412-419. [DOD] [CrossRef]
- Ding K, Zhou K, He F, Shen Y. LDA - A java-based linkage disequilibrium analyzer. Bioinformatics 2003. 19:2147-2148. [DOD] [CrossRef]
- Bland JM, Altman DG. The odds ratio. BMJ 2000. 320:1468. [DOD] [CrossRef]
- Damcott CM, Ott SH, Pollin TI, Reihart LJ, Wang J, O'Connell JR, Mitchell BD, Shuldiner AR. Genetic variation in adiponectin receptor 1 and adiponectin receptor 2 is associated with type 2 diabetes in the Old Order Amish. Diabetes 2005. 54:2245-2250. [DOD] [CrossRef]
- Vaxillaire M, Dechaume A, Vasseur-Delannoy V, Lahmidi S, Vatin V, Lepretre F, Boutin P, Hercberg S, Charpentier G, Dina C, et al. Genetic analysis of ADIPOR1 and ADIPOR2 candidate polymorphisms for type 2 diabetes in the Caucasian population. Diabetes 2006. 55:856-861. [DOD] [CrossRef]
- Richardson DK, Schneider J, Fourcaudot MJ, Rodriguez LM, Arya R, Dyer TD, Almasy L, Blangero J, Stern MP, DeFronzo RA, et al. Association between variants in the genes for adiponectin and its receptors with insulin resistance syndrome (IRS)-related phenotypes in Mexican Americans. Diabetologia 2006. 49:2317-2328. [DOD] [CrossRef]
- Broedl UC, Lehrke M, Fleischer-Brielmaier E, Tietz AB, Nagel JM, Goke B, Lohse P, Parhofer KG. Genetic variants of adiponectin receptor 2 are associated with increased adiponectin levels and decreased triglyceride/VLDL levels in patients with metabolic syndrome. Cardiovasc Diabetol 2006. 5:11. [DOD] [CrossRef]
- Tirosh A, Shai I, Tekes-Manova D, Israeli E, Pereg D, Shochat T, Kochba I, Rudich A. Normal fasting plasma glucose levels and type 2 diabetes in young men. N Engl J Med 2005. 353:1454-1462. [DOD] [CrossRef]
- Trujillo ME, Scherer PE. Adiponectin - journey from an adipocyte secretory protein to biomarker of the metabolic syndrome. J Intern Med 2005. 257:167-175. [DOD] [CrossRef]
- Yamauchi T, Kamon J, Minokoshi Y. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 2002. 8:1288-1295. [DOD] [CrossRef]
- Combs TP, Berg AH, Obici S, Scherer PE, Rossetti L. Endogenous glucose production is inhibited by the adipose derived protein Acrp30. J Clin Invest 2001. 108:1875-1881. [DOD]
- Ravussin E, Smith SR. Increased fat intake, impaired fat oxidation, and failure of fat cell proliferation result in ectopic fat storage, insulin resistance, and type 2 diabetes mellitus. Ann N Y Acad Sci 2002. 967:363-378. [DOD]
- Liu YM, Michael MD, Kash S, Bensch WR, Monia BP, Murray SF, Otto KA, Syed SK, Bhanot S, Sloop KW, et al. Deficiency of adiponectin receptor 2 reduces diet-induced insulin resistance but promotes type 2 diabetes. Endocrinology 2007. 148:683-692. [DOD] [CrossRef]
This article has been cited by other articles:

|
Gender-specific differences in the association of adiponectin gene polymorphisms with body mass index
Tabatabaei-Malazy O, Hasani-Ranjbar S, Amoli MM, Heshmat R, Sajadi M, Derakhshan R, Amiri P, Namakchian M, Rezazadeh E, Tavakkoly-Bazzaz J, Keshtkar A, Larijani B
Rev Diabet Stud 2010. 7(3):241-246
|
|

|
Analysis of Polymorphisms at the Adiponectin Gene Locus in Association with Type 2 Diabetes, Body Mass Index and Cardiovascular Traits in Latvian Population
Kalnina I, Latkovskis G, Nkitina-Zake L, Mackevics V, Raitis Peculis R, Kapa I, Fridmanis D, Erglis A, Pirags V, Klovins J
Proc Latvian Acad Sci 2009. 63(4-5):174-179
|
|
|