Original Data
| Rev Diabet Stud,
2017,
14(2-3):311-328 |
DOI 10.1900/RDS.2017.14.311 |
Variations in ADIPOR1 But Not ADIPOR2 are Associated With Hypertriglyceridemia and Diabetes in an Admixed Latin American Population
Gustavo Mora-García1, María S. Ruiz-Díaz1, Fabian Espitia-Almeida2, Doris Gómez-Camargo1
1Doctorate in Tropical Medicine, Faculty of Medicine, Universidad de Cartagena. Cartagena de Indias, Colombia
2Biochemistry Master Program, Faculty of Medicine, Universidad de Cartagena. Cartagena de Indias, Colombia
Address correspondence to: Gustavo Mora-García, e-mail: gmorag@unicartagena.edu.co
Manuscript submitted May 3, 2017; resubmitted July 17, 2017; accepted August 29, 2017.
Keywords: diabetes mellitus, adiponectin, adiponectin receptor, ADIPOR1, ADIPOR2, hypertriglyceridemia, genetic association study
Abstract
BACKGROUND: Adiponectin is a hormone secreted by adipose tissue. It regulates glycolysis and lipolysis and is involved in the pathophysiology of diabetes and related disorders. Its activity is mainly mediated by the transmembrane receptors AdipoR1 and AdipoR2, which are encoded by ADIPOR1 (1q32.1) and ADIPOR2 (12p13.33) genes, respectively. In genetic association studies, single nucleotide polymorphisms (SNPs) in or near these genes have been associated with metabolic alterations. However, these relationships are still controversial. AIM: The aim of this work was to analyze possible associations between ADIPOR1/2 and diabetes and other metabolic disorders. METHODS: A genetic association study was carried out in an admixed Latin American population. A sample of 200 adults was analyzed. Clinical and serum-biochemical characteristics were measured to diagnose obesity, abdominal obesity, hypertension, hyperglycemia, hypertriglyceridemia, low HDLc, insulin resistance (HOMA-IR), and diabetes. Three SNPs were genotyped in ADIPOR1 (rs10494839, rs12733285, and rs2275737) and ADIPOR2 (rs11061937, rs11612383, and rs2286383). For the association analysis, an additive model was assessed through logistic regression. An admixture adjustment was performed using a Monte-Carlo-Markov-Chain method, assuming a three-hybrid substructure (k = 3). RESULTS: Two SNPs in ADIPOR1 were associated with diabetes: rs10494839 (OR = 3.88, adjusted p < 0.03) and rs12733285 (OR = 4.72, adjusted p < 0.03). Additionally, rs10494839 was associated with hypertriglyceridemia (OR = 2.16, adjusted p < 0.01). None of the SNPs in ADIPOR2 were associated with metabolic disorders. CONCLUSIONS: ADIPOR1 was consistently associated with diabetes and hypertriglyceridemia. This association was maintained even after adjusting for genetic stratification. There were no significant associations involving ADIPOR2.
Abbreviations: ADIPOR1 - adiponectin receptor 1 (gene); ADIPOR2 - adiponectin receptor 2 (gene); AdipoR1 - adiponectin receptor 1; AdipoR2 - adiponectin receptor 2; AIM - ancestry informative marker; ANOVA - analysis of variance; BMI - body mass index; Fis - Wright's coefficient of inbreeding; GPAQ - Global Physical Activity Questionnaire; HbA1c - glycosylated hemoglobin A1c; HDLc - high-density lipoprotein cholesterol; HOMA-IR - homeostasis model assessment for insulin resistance; IR - interquartile range; JNC8 - 8th Joint National Committee; MAF - minor allele frequency; NOS - nitric oxide synthase; PAQR - progestin and AdipoQ receptor; qPCR - quantitative polymerase chain reaction; r2 - correlation coefficient; SNiPA - single nucleotide polymorphism annotator; SNP - single nucleotide polymorphism; T2DM - type 2 diabetes mellitus; WHO - World Health Organization; Y-STR - Y chromosome short tandem repeats
1. Introduction
Adiponectin is a 30 kDa collagen-like protein with autocrine, paracrine, and endocrine activity, mainly secreted by adipocytes; it has physiologic effects on insulin sensitivity and energetic homeostasis [1, 2]. Most systemic responses to adiponectin are mediated by two specific transmembrane receptors known as AdipoR1 and AdipoR2, which are members of the progesterone and AdipoQ receptor (PAQR) family. They are abundantly expressed in many cellular types, such as hepatocytes, endothelial cells, myocytes, and neurons, among others [3, 4].
Adiponectin receptors have a seven-trans-membrane-domain architecture that confers their ligand recognition function on the extracellular surface; it also has a zinc-binding site and an intrinsic ceramidase activity oriented to the intracellular space [3, 5]. The appropriate stimulus triggers conformational changes in the receptor structure that increases the intrinsic enzymatic activity. In muscle cells, this activity enhances the beta-oxidative pathway, glucose capture, and glycolysis, whereas in hepatocytes, these changes reduce gluconeogenesis [6-9]. Other responses have linked AdipoR1/R2 to the satiety cycle in the hypothalamus, NOS-induced vascular relaxation in endothelial cells, and inflammatory cascades in macrophages, among others [10-12].
It has been widely proposed that variations in genes encoding these receptors may be associated with alterations in some biological processes in which the hormone is involved. It has been found that several single nucleotide polymorphisms (SNPs) in ADIPOR1 (1q32.1) and ADIPOR2 (12p13.33) are associated with plasma concentrations of adiponectin, abdominal obesity, and related disorders (i.e., insulin resistance, hyperglycemia, and high serum triglycerides, among others) [13-16]. According to these data, both genes have been raised as promising targets of treatment approaches to induce complex metabolic alteration. However, some controversial issues remain based on reports of contradictory evidence where no significant associations were found [17, 18].
The Latin American population has recently been exposed to increased racial admixture. Phenotypic expressions of metabolic traits are highly diverse in this population, and the prevalence of related disorders shows remarkable variation between countries [19, 20]. This diversity is an opportunity to uncover associated genetic factors that might be helpful to solve current discrepancies [21]. On this basis, we conducted a study aiming to describe the relationship between common variations in ADIPOR1/R2 with metabolic disorders and syndromes in a Latin America population.
2. Methods and materials
2.1 Subjects
A cross-sectional study was carried out in Cartagena de Indias, a city of nearly 1 million inhabitants located at the Colombian Caribbean Coast [22]. The genetic stratification of this population has been described elsewhere; it is known to be a three-hybrid admixed population including European (60%), African (30%), and Amerindian (10%) ancestry [23, 24]. A sample of 200 adults was employed to achieve 80% power for discrimination of a genetic association with a 2.5/3.5 (heterozygotes/homozygotes) odds ratio, assuming an outcome with 25% prevalence, 25% minor allele frequency, complete linkage disequilibrium (D' = 1), and 5% alpha-coefficient (for type I error), according to calculations described by other authors. We used the browser program Genetic Power Calculator (http://zzz.bwh.harvard.edu/gpc/cc2.html) [25, 26].
Subjects were selected from the urban zone and only no-sibling adults (18-80 years) were allowed to participate. To identify possible cases of consanguinity, individuals with similar surnames were contacted by telephone to discard family relations. On this basis, first- and second-degree siblings were excluded, retaining only one of the subjects for further analysis. Individuals with a personal history of primary endocrine disorders, genetic disease, or surgical treatment for obesity were excluded. Also, pregnant or breast feeding women were excluded. All subjects were asked to participate, and their written informed consent was obtained, following the Universidad de Cartagena ethics committee recommendations.
Selected subjects were enrolled by a trained physician, and underwent a medical examination focused on sociodemographic variables and clinical history of metabolic disorders. Physical activity and sedentary behavior were noted in the Global Physical Activity Questionnaire (GPAQ) developed by the World Health Organization (WHO) for physical activity surveillance [27].
2.2 Anthropometric data
Anthropometric parameters (height, weight, and waist circumference,) were measured during physical examination following the international diabetes guidelines for the metabolic syndrome and WHO recommendations [28-30]. Height was measured to the nearest half centimeter using a stadiometer with the participant barefoot, and registered in meters. Weight was measured to the nearest 0.1 kilogram (kg) using a calibrated digital scale, with the subjects wearing light clothes without shoes. Body mass index (BMI) was calculated as weight (kg) divided by the square of height (m2). Using an inelastic tape measure, waist circumference was measured at two centimeters below the umbilicus with the subjects in a standing position, their weight equally distributed on both feet, arms at their sides, and head facing straight forward at the end of a normal expiration, ensuring that the tape did not compress the skin and was parallel to the floor. Blood pressure was measured using a sphygmomanometer after a resting period of at least 5 min, using the auscultatory method according to recommendations from the 8th Joint National Committee (JNC8) [29].
2.3 Blood samples
A whole-blood sample was collected under 8-hour fasting conditions to measure serum concentrations of glucose, triglycerides, HDL cholesterol (HDLc), insulin, adiponectin, and the proportion of glycosylated hemoglobin (HbA1c). An aliquot blood was stored for further genetic analyses.
2.4 Definition of obesity, hypertension, and the metabolic syndrome
Using the anthropometric parameters and biochemical data, metabolic traits (BMI) were defined following the criteria defined by the WHO:
- Normal weight: BMI 18.51-24.99 kg/m2
- Overweight: BMI 25-29.99 kg/m2
- Obesity as BMI ≥ 30 kg/m2
Body weight excess was determined by the addition of overweight and obesity categories [31].
Hypertension was defined according to the JNC8 [29]:
- Younger than 60 years old: systolic ≥140 mmHg, diastolic ≥90 mmHg
- Adults 60 years old or more: systolic ≥150 mmHg, diastolic ≥90 mmHg.
The metabolic syndrome and its related alterations were defined according to Joint Interim Statement criteria:
- Abdominal obesity: waist circumference men ≥90 cm, women ≥80 cm.
- Dyslipidemia: hypertrigliceridemia: TG ≥150 mg/dl (1.7 mmol/l) or drug treatment for high serum triglycerides. Low HDLc: men, HDLc <40 mg/dl (1.0 mmol/l); women, HDLc <50 mg/dl (1.3 mmol/l)).
- Hyperglycemia: glucose impaired fasting ≥100 mg/dl or drug treatment for elevated glucose [32].
2.5 Definition of insulin resistance
The definition of insulin resistance was based on the homeostasis model assessment for insulin resistance (HOMA-IR), as described by other authors. Subjects in the last quintile were considered to be insulin resistant [33]. The same procedure was applied to determine high serum adiponectin status in the sample population. Type 2 diabetes mellitus (T2DM) was defined by HbA1c ≥ 6.5%, as proposed by the American Diabetes Association, or by a history of the disease [34].
2.6 Genotyping
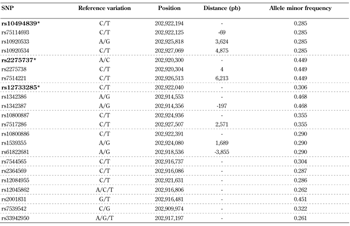
For genotyping assays prior to molecular procedures, common variants in ADIPOR1 and ADIPOR2 were identified using 1000 genome project reports, with the bioinformatic resource SNiPA [35, 36]. SNPs with a minor allele frequency (MAF) ≥0.25 and correlation coefficient (r2) = 1.0 were selected using data from populations with European ancestry (Tables A1 and A2). Among the tagged variants, a set of SNPs in both genes was picked. For ADIPOR1, the included variants were:
- rs10494839 (proxy for rs75114693, rs10920533, rs10920534, rs10920537, rs6666089, and rs2232853)
- rs12733285
- rs2275737 (proxy for rs2275736, rs2275738, and rs7514221)
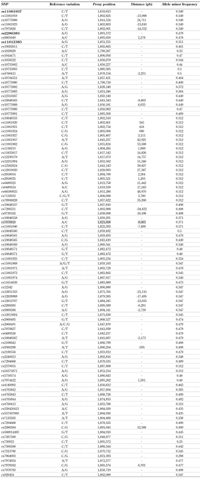
Similarly, for ADIPOR2 the variants were:
- rs11061937 (proxy for rs11061919, rs11061923, 10773986, 7975826)
- rs11612383
- rs2286383 (proxy for rs9805049, rs2286384)
These SNPs are proxy markers for several variants identified through datasets from the 1000 genome project; they include positive antecedents of genetic association with metabolic traits and related diseases identified previously [37-41].
Selected SNPs were genotyped with quantitative polymerase chain reaction (qPCR) using specific TaqMan probes (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Allelic discrimination was performed automatically with endpoint fluorescent data, which were analyzed using StepOne Real-Time PCR Software (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
2.7 Statistics
For statistical procedures, sociodemographic data, personal and family antecedents, as well as anthropometric measures and serum concentrations were described using main tendency and frequency values. When appropriate, mean values were compared using Student's t-test, and frequencies were compared with X2 or Fisher's exact tests.
Allelic and genotypic frequencies were determined by direct count, linkage disequilibrium was estimated employing Arlequin 3.5 [42], and Hardy-Weinberg Equilibrium was assessed through Fis values using Genetix 4.05 software. Given that ADIPOR1 and ADIPOR2 represent distant loci, genetic associations were analyzed separately. Associations between continuous traits and genotype distributions were analyzed using the Kruskal-Wallis test and analysis of variance (ANOVA). Relations between categorical outcomes (i.e., hypertension or T2DM) and genotypes were determined with X2 tests. In both cases, recessive and dominant models were assessed using null hypothesis tests [43]. Additive models for genotype-phenotype associations were evaluated through logistic regression, where risk alleles were equal to the unit (risk allele = 1), and anthropometric alterations and metabolic disorders were interpreted as categorical outcomes. Logistic regression was adjusted by age, sex, and BMI, except for body weight excess (overweight and obesity) analysis, for which only age and sex were used. These procedures were performed with R version 3.2.1 and the package PredictABEL 1.2-2 [44, 45]. Based on Bonferroni's correction for multiple testing, p-values <0.017 were considered as statistically significant.
Associations were adjusted by genetic stratification assuming a three-hybrid substructure (k = 3) using a Bayesian approach (Markov Chain Monte Carlo, or MCMC) with 100,000 replications using the STRAT software version 1.1 for DOS/Windows [46].
2.8 Admixture pattern
The three-hybrid admixture pattern applied is based on previous reports from Cartagena de Indias, where ancestry informative markers (AIMs) and the Y chromosome were used to describe local genetic substructure and ancestry distribution [23, 24, 47, 48]. In this study, a total of 17 Y chromosome short tandem repeats (Y-STR; AmpFLSTR® Yfiler® PCR Amplification Kit, Thermo Fisher, Inc., USA) were used to confirm the admixture in sampled males (data not shown).
3. Results
3.1 Age, income, and physical activity – descriptive data
A total of 200 subjects were included in the study (44.5% men and 55.5% women), with an average age of 33.7 ± 14.2 years. Age ranges were distributed according to a population pyramid [49]:
- 18-29 years = 53.8%
- 30-39 years = 17.1%
- 40-49 years = 13.6%
- 50-59 years = 10.6%
- 60-69 years = 2.5%
- 70 or more years = 2.5%
Given that economic status is highly correlated with nutritional conditions, total family income was registered for all subjects. The median income was US$ 364 per month interquartile range (IR, $241-$690).
Sedentary behavior was the most common pattern identified among sampled subjects, with 540 min/day IR (360-720) of low energy-expenditure activities as a median tendency. Mild-intensity activities were regularly executed by 38 (19%) subjects with a median of 60 min/day IR (30-120), and 40 (20%) subjects were found to perform high-intensity activities on a weekly basis with a median of 60 min/day IR (56-120).
3.2 Anthropometric data and diabetes incidence
Regarding anthropometric parameters, median values for weight, height, and BMI were 68.7 kg IR (56.85-78.45), 165 cm (158.3-172), and 24.9 kg/m2 (21.9-27.68), respectively. The mean value for waist circumference was 88.1 ± 13.5 cm. The median values for blood pressure were 110 mmHg IR (100-118) (systolic) and 88.0 mmHg IR (79.85-97.38) (diastolic). Serum concentration of glucose was 90.0 mg/dl IR (86-96) (5.0 mmol/liter IR (4.8-5.3)), triglycerides 173.0 mmol/l IR (156-192) (2.0 mmol/l IR (1.8-2.2)), HDLc 48.0 mg/dl IR (45-51) (1.2 mmol/l IR (1.2-1.3)), insulin 10 µUI/ml (10-11.65) (71.8 mmol/l IR (71.8-83.6)), and adiponectin 15.5 ng/ml IR (12.5-18-95). The average proportion of HbA1c was 5.0% IR (4.2-6.0). The mean value of HOMA-IR was estimated 2.3 IR (2.1-3.0).
Overweight and obesity were identified in 48.2% and abdominal obesity in 65.3% of individuals. The frequency of hypertension was 24.1%, whereas hyperglycemia, hypertriglyceridemia, and low HDLc were found in 19.1%, 81.9%, and 37.7%, respectively. According to personal antecedents and HbA1C values, T2DM was present in 4% of the sampled subjects. The metabolic syndrome was diagnosed in 85 (42.7%) subjects, and among these, five criteria were met by 5 subjects (2.5%), four by 21 (10.6%), and three by 59 (29.6%).
3.3 Genotyping
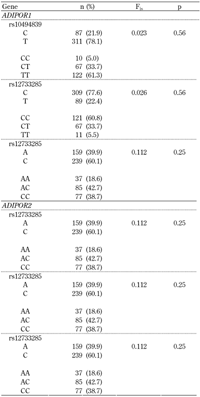
Genotyping assays were possible for 199 subjects. In the ADIPOR1 gene:
- rs10494839 showed an MAF (C allele) of 21.9% (n = 87)
- rs12733285 MAF (T allele) was 22.4% (n = 89)
- rs2275737 MAF (A allele) was 39.9% (n = 159)
In ADIPOR2, the MAF for the three assessed SNPs were:
rs11061937 (C allele) 39.7% (n = 158)
rs11612383 (A allele) 33.9% (n = 135)
rs2286383 (A allele) 35.7% (n = 142)
Genotype distributions and Fis values for Hardy-Weinberg equilibrium are shown in Table 1.
Table
1.
Allelic frequencies and genotype distributions for ADIPOR1 and ADIPOR2 genes |
|
 |
 |
3.4 Genetic associations of continuous anthropometric and metabolic variables
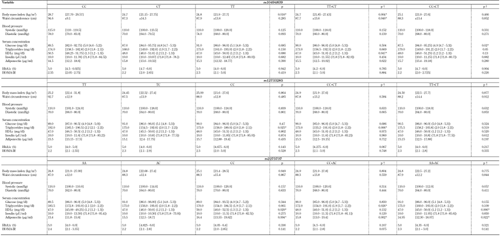
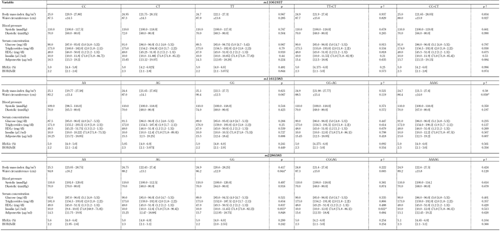
Median and mean values of anthropometric and metabolic variables were compared according to genotype distributions. In ADIPOR1, no differences were found when the rs12733285 and rs2275737 genotypes were applied as classification criteria. However, differences in BMI among the three genotypes were found in rs10494839 (p < 0.02). Also, the TT+CT group had lower BMI (p < 0.01), lower waist circumference (p < 0.05), and lower HDLc (p < 0.05) than the CC homozygotes; CC+CT genotypes had lower glucose values than TT homozygotes (p < 0.03; Table A3). Similarly, in rs12733285, TT homozygotes had higher triglyceride concentrations than CC homozygotes, CT heterozygotes (p < 0.03), and CC+CT genotypes (p < 0.01). In rs2275737, the CC+AC group had a lower triglyceride value than AA homozygotes, and there were differences in mean HDLc and adiponectin levels between the three genotypes (p < 0.03; Table A3).
Furthermore, AA+AC genotypes had lower HDLc levels than CC homozygotes (p < 0.01), CC+AC genotypes had higher levels of adiponectin than AA homozygotes (p < 0.01), and AA+AC genotypes had lower levels of adiponectin than CC homozygotes (Table A3). For ADIPOR2, the AA+AG group had lower values for waist circumference than GG homozygotes in rs11612383 (p < 0.04). Additionally, differences in waist circumference and insulin values between rs2286383 genotypes were found (p < 0.05; Table A4).
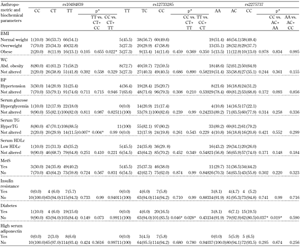
3.5 Genetic associations of metabolic disorders
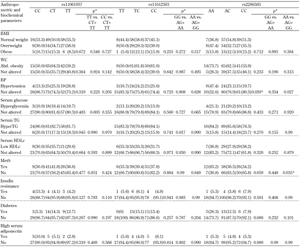
We also compared the frequencies of anthropometric parameters with metabolic disorders. We found differences for ADIPOR1 among cases for high serum triglycerides, with differences in the genotype distribution for rs10494839 (p < 0.01). A recessive model showed a higher occurrence of hypertriglyceridemia in TT homozygotes for rs10494839 when compared with CC+CT genotypes (p < 0.01) (Table 2). Similarly, T2DM cases were more frequent among CC homozygotes for rs12733285 (p < 0.05) and for rs2275737 (p < 0.05). These observations were corroborated by a recessive model (Table 2). In ADIPOR2, high blood pressure was more frequent in subjects with the AA genotype for rs2286383 (p < 0.05), and the metabolic syndrome was less frequent among AA homozygotes (p < 0.04; Table 3).
Table
2.
Genotype distributions for ADIPOR1 single nucleotide polymorphisms according to metabolic traits |
|
 |
 |
Legend:
Data are numbers (%). * p < 0.05, Abbreviations: Abd. obesity - abdominal obesity, BMI - body mass index, HDLc - high-density lipoprotein cholesterol, HyperTG - hypertriglyceridemia, MetS - metabolic syndrome, TG - triglycerides, WC - waist cricumference. |
|
Table
3.
Genotype distributions for ADIPOR2 single nucleotide polymorphisms according to metabolic traits |
|
 |
 |
Legend:
Data are numbers (%). * p < 0.05, Abbreviations: Abd. obesity - abdominal obesity, BMI - body mass index, HDLc - high-density lipoprotein cholesterol, HyperTG - hypertriglyceridemia, MetS - metabolic syndrome, TG - triglycerides, WC - waist cricumference. |
|
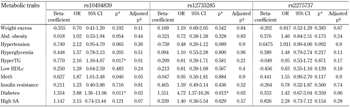
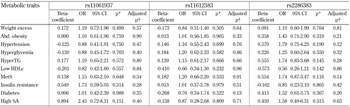
The assessment of an additive model revealed significant associations between rs10494839 (ADIPOR1), high serum triglycerides (OR = 2.16, p = 0.01) and T2DM (OR = 3.88, p = 0.01), in which the T allele was identified as a risk factor. The statistical significance of these relationships persisted after admixture adjustment for a three-hybrid population (p = 0.03). Moreover, rs12733285 (ADIPOR1) was found to be associated with an increased risk of T2DM (OR = 4.72, p = 0.01). This finding remained statistically significant when the admixture adjustment was applied (p = 0.03; Table 4). None of the SNPs analyzed in ADIPOR2 was associated with any of the metabolic traits included in this study (Table 5).
Table
4.
Association of ADIPOR1 gene variants with metabolic disorders |
|
 |
 |
Legend:
*p < 0.017 were considered as statistically significant (Bonferroni correction). † Admixture adjustment with Monte-Carlo-Markov Chain (k = 3). An additive model was assessed through a logistic regression where risk alleles were interpreted as the unit. Genetic variations in rs10494839 (CC = 0, CT = 1, and TT = 2), rs12733285 (TT = 0, CT = 1, and CC = 2), and rs2275737 (AA = 0, AC = 1, and CC = 2) were included as independent variables. Age, sex, and body mass index were included as confounding variables (except for the analysis of body weight excess in which only age and sex were used). An admixture adjustment was performed through a Monte-Carlo-Markov Chain where a three-hybrid genetic stratification was assumed. Abbreviations: Abd. obesity - abdominal obesity, HDLc - high-density lipoprotein cholesterol, HyperTG - hypertriglyceridemia, MetS - metabolic syndrome, SA - serum adiponectin. |
|
Table
5.
Association of ADIPOR2 gene variants with metabolic disorders |
|
 |
 |
Legend:
*p < 0.017 were considered as statistically significant (Bonferroni correction). † Admixture adjustment with Monte-Carlo-Markov Chain (k = 3). An additive model was assessed through a logistic regression where risk alleles were interpreted as the unit. Genetic variations in rs11061937 (TT = 0, CT = 1, and CC = 2), rs11612383 (GG = 0, AG = 1, and AA = 2), and rs2286383 (GG = 0, AG = 1, and AA = 2) were included as independent variables. Age, sex, and body mass index were included as confounding variables (except for the analysis of body weight excess in which only age and sex were used). An admixture adjustment was performed through a Monte-Carlo-Markov Chain where a three-hybrid genetic stratification was assumed. Abbreviations: Abd. obesity - abdominal obesity, HDLc - high-density lipoprotein cholesterol, HyperTG - hypertriglyceridemia, MetS - metabolic syndrome, SA - serum adiponectin. |
|
4. Discussion and conclusions
In admixed populations, understanding the genotype/phenotype interactions behind complex diseases remains a challenging task because genetic stratification is a source of false positives caused by confounding phenomena [50]. Therefore, appropriate analysis requires the application of rigorous ancestry-adjusted statistical procedures, and high-powered methodological designs are often needed to uncover common associations [46, 50]. Despite this challenge, in the present study we found consistent evidence for associations between ADIPOR1 with high serum triglycerides and T2DM in a Latin American population, where a three-hybrid substructure has been previously reported [23, 24, 47, 48].
The SNPs rs10494839 and rs12733285, located in ADIPOR1, were found to be associated with T2DM, which represents an important finding of the present research, considering that this relationship has often been elusive in genetic association studies. According to Bermudez et al. (2013), there is little evidence describing an association between ADIPOR1/2 and T2DM in human populations [51]. In fact, Peters et al. (2013) found that ADIPOQ but not ADIPOR1/2 was associated with T2DM, and Kim et al. (2009) reported a lack of significant associations of ADIPOR1/2 genes with this disease [18, 52]. On the other hand, Mather et al. (2012) found two SNPs (rs1342387 and rs12733285) in ADIPOR1 to be significantly associated with diabetes incidence [41], which strongly supports the results of the current study. Similarly, Jin et al. (2014) found an association between two SNPs in ADIPOR1 (rs3737884 and rs16850797) and T2DM in a Chinese population [53], and recently another study successfully replicated these findings with subjects from Northeastern China. Moreover, a third SNP (rs7514221) in ADIPOR1 was also found to be associated with an increased risk of T2DM [54].
Adiponectin receptors have been widely reported to be involved in the development of T2DM, in both in vitro and in vivo studies, where the regulatory activity of adiponectin signaling controlling beta-oxidative and glycolytic pathways has been demonstrated [6, 55]. In human populations, variations in the physiologic response to adiponectin have been related to insulin resistance and T2DM. Therefore, both ligand and receptor are thought to be promising targets for novel therapeutics [56]. In this regard, an agonist of adiponectin receptors (AdipoRon) has been reported to ameliorate T2DM in rodents [57], and recently, it was suggested that most of its pharmacological effects are mediated through AdipoR1, although details of intracellular signaling triggered by this molecule remain unclear [58]. In this connection, our results encourage analyses of possible effects of genetic variations in ADIPOR1 on receptor expression and cellular responses in relation to available agonists because a pharmaco-genomic interaction would be an interfering factor for novel therapies.
Insulin resistance has been described as an early stage in the pathogenesis of glucose intolerance and T2DM [59]. Therefore, we assessed a possible relationship between ADIPOR1/2 variations, serum insulin concentrations, and HOMA-IR in the present study. Despite the cumulative biologic and epidemiologic evidence regarding this relationship, none of the analyzed SNPs was associated with insulinemia, HOMA-IR values, or insulin resistance. Other authors have obtained similar results, reporting that no genotype/phenotype associations were found when a set of SNPs covering adiponectin receptors genes were analyzed [18]. Rasmussen-Torvik et al. (2009) genotyped the same three variations in ADIPOR1 that were selected in the present work (rs10494839, rs12733285, and rs2275737), and they found no significant relationship with insulin resistance. However, in that study, another SNP in ADIPOR1 (rs1342387) was associated with an increased diabetes risk [60]. It is well known that adiponectin activity progressively decays, while conversely, serum concentrations of adiponectin increase during the transition from insulin-resistance to T2DM, until a so-called "adiponectin-resistance" status is reached [61, 62]. It is therefore possible that groups with altered responses to insulin represent a diverse population of subjects with different levels of adiponectin activity, which may attenuate the effects of genetic variations on receptor function, causing a confounding scenario for association analysis.
Hypertriglyceridemia was found to be associated with the SNP rs10494839 (ADIPOR1) both in the regression model and the MCMC (Table 4). To the best of our knowledge, there is scarce evidence regarding this genotype/phenotype relationship, and there have been no previous reports where this particular SNP has been found to be involved as a risk factor for high serum triglycerides. The closest precedents were published by Jin et al. (2014) who found that rs16850797 and rs3737884 SNPs in ADIPOR1 were associated with higher triglyceride levels in subjects with T2DM and coronary artery disease [53], and by Potapov et al. (2008) who also found higher serum triglyceride concentrations related to rs2275738 (ADIPOR1) among subjects with T2DM, but not in the control group [40]. In contrast, Ferguson et al. (2010) genotyped three ADIPOR1 SNPs (rs2275737, rs10753929, and rs10920533), and found that none of them were related to serum levels of triacylglycerol. In a study carried out with Finnish and Swedish adults, the SNP rs6666089 in ADIPOR1 also had no association with triacylglycerol levels [63]. Additionally, Yeh et al. (2008) found no association between ADIPOR1 and serum triglycerides in a multi-ethnic Brazilian sample where two variants in regulatory regions were genotyped, which represents one of the most relevant antecedents for South American populations [64].
To date, most findings have pointed to ADIPOR2 variants as a genetic factor related to high serum triglycerides. Richardson et al. (2006) found 14 variants in ADIPOR2, but none in ADIPOR1, related to serum triglycerides in a study where 6 SNPs in ADIPOR1 and 24 SNPs in ADIPOR2 were analyzed [65]. Moreover, Broedl et al. (2006) found a cluster of three SNPs in ADIPOR2 that were related to lower levels of triglycerides in a population sample of European ancestry [16], and Potapov et al. (2008) found that TT homozygotes in rs11061971 (ADIPOR2) have higher serum triglyceride concentrations [40]. In the study carried out by Kotronen et al. (2009), another SNP in ADIPOR2 (rs767870) was associated with increased levels of triglycerides in men without lipid-lowering medication [63]. Recently, Castilhos et al. (2015) found two ADIPOR2 SNPs (rs11061925 and rs929434), but none of the two in ADIPOR1 were associated with serum triglycerides in men infected with HIV under anti-retroviral treatment [66].
Most authors have suggested that the molecular mechanism behind these genetic associations with serum triglycerides is related to the metabolic effects of adiponectin that increase fatty acid oxidation through AMPK and APPL1 activation [67]. Activation of these pathways leads to enhanced lipolysis and reduced lipid metabolites, which interfere with intracellular insulin signaling [68], favoring insulin sensitivity, and preventing the development of T2DM. Therefore, it is possible that the results from the current study that involve T2DM and hypertriglyceridemia indicate a single phenomenon instead of two independent genotype/phenotype relationships. To solve this issue, a novel logistic regression model adjusted by T2DM prevalence (as well as sex and age) was performed, and the association between rs10494839 and high serum triglycerides remained statistically significant (p = 0.013). Additionally, when subjects with T2DM were excluded, this relationship also remained statistically significant (p = 0.026). Therefore, these findings support further studies focused on the potential role of AdipoR1 and adiponectin in the treatment of high serum triglycerides. It is rational to approach the effects of pharmacologic stimuli on liposome and lipid droplet formation in humans.
Another remarkable finding involving rs2275737 indicates that HDLc concentrations were lower in subjects with the AA genotype and also in the group with AA+AC genotypes (p < 0.01; Table A3). Regarding this relationship, Jin et al. (2014) found previously that subjects with T2DM and risk genotypes (GG+GA) in rs3737884 had lower concentrations of HDLc in a genetic association study [53]. Similarly, Yeh et al. (2008) reported that a variant in the regulatory region of ADIPOR1 is associated with lower HDLc in Brazilian subjects with African ancestry [64].
Although adiponectin level medians were associated with the rs2275737 SNP (Table A3), no associations between a metabolic trait and plasma levels of adiponectin were found. In contrast, several authors have described associations:
- Chen et al. (2017) showed that Adiponectin is inversely associated with the metabolic syndrome and high arterial stiffness [69].
- Some population studies have revealed a correlation between plasma levels of adiponectin and HDLc, as was observed by Medina-Urrutia et al. (2015), who showed that low adiponectin levels are closely related to low serum HDLc in adults with Mexican-Mestizo ancestry [70].
- In Japanese individuals, Matsushita et al. (2014) found a similar phenomenon [71], and Hanley et al. (2007) showed that adiponectin levels are positively associated with HDLc in Hispanics and African-Americans [72].
As expected, anti-dyslipidemic therapy has been observed to influence adiponectin secretion. Statins and fibrates have been generally found to increase adiponectin levels in groups under medical treatment [73]. Thus, it is reasonable to suspect a biological interaction between these genetic variations and the adiponectin release induced by pharmacological activity of statins and fibrates, which may be a research focus in future studies.
Despite the statistically significant results, there are some limitations that should be considered. Firstly, the sample size in the current study was small, which may be a confounding issue in proposing definitive genotype/phenotype relationships. Nonetheless, it was possible to identify consistent genetic associations, even after application of admixture-adjustment methods, suggesting that sampling strategies could have corrected related bias. Therefore, an expansion of the present sample is strongly encouraged to obtain results from a study with greater predictive power. Secondly, an elevated proportion of the current sample had hypertriglyceridemia; this finding could be a consequence of selection bias. Therefore, further case-control studies would be helpful to verify the actual nature of the genetic relationships described in this study. As a matter of fact, an expanded sample (n = 1,500) of the same population has been analyzed, and similar frequencies of high serum triglycerides and other metabolic alterations have been found (awaiting publication), suggesting that acquired dyslipidemia may be an issue of particular relevance in the population.
In summary, according to the results of the present study and previous data, consistent evidence has been found for an association between genetic variations in ADIPOR1 with high serum triglycerides and T2DM. These findings encourage future studies on potential biological and pharmacogenomic interactions.
Disclosures: This work was carried out with the financial support of the Universidad de Cartagena, Cartagena de Indias, (Vicerrectoría de Investigaciones, Séptima convocatoria para la financiación de proyectos de investigación (Res 04379 de 2014)) Colombia. M.S. Ruiz-Diaz and F. Espitia-Almeida were supported by the Colombian Administrative Department of Science, Technology, and Innovation (COLCIENCIAS) by resolution no. 2286. G. Mora-Garcia was supported by COLCIENCIAS, grant no. 528-2012, for doctoral fellowship.
Appendix
Table
A1.
Single nucleotide polymorphisms selected in the adiponectin receptor 1 gene (ADIPOR1) employing data from the 1000 genome project |
|
 |
 |
Legend:
A total of 22 SNPs were observed in ADIPOR1, using the bioinformatic resource SNiPA. Several of the variants were found in linkage disequilibrium (LD) with each other taking into account a correlation coefficient (r2) = 1.0. The dotted lines denote the groups of SNPs in LD. Two of the genotyped SNPs were in LD with five of the 22 variants: rs10494839 with three (rs75114693, rs10920533, rs10920534) and rs227557 with two (rs227538, rs7514221). For this selection, a minor allele frequency > 0.25 was used. *Genotyped SNPs. |
|
Table
A2.
Single nucleotide polymorphisms selected in the adiponectin receptor 2 gene (ADIPOR2) employing data from the 1000 genome project |
|
 |
 |
Legend:
A total of 101 SNPs were observed in ADIPOR2, using the bioinformatic resource SNiPA. Several of the variants were found in linkage disequilibrium (LD) with each other taking into account a correlation coefficient (r2) = 1.0. The dotted lines denote the groups of SNPs in LD. Five of the genotyped SNPs were in LD with five of the 101 variants: rs11061937 with four (rs11061919, rs10773986, rs11061923, rs7975826) and rs2286383 with one (rs9805049). For this selection, a minor allele frequency > 0.25 was used. *Genotyped SNPs. |
|
Table
A3.
Mean values of anthropometric and biochemistry parameters according to the genotype for ADIPOR1 SNPs |
|
 |
 |
Legend:
Data are median (1st quartile - 3rd quartile), mean ± SD. |
|
Table
A4.
Mean values of anthropometric and biochemistry parameters according to the genotype for ADIPOR2 SNPs |
|
 |
 |
Legend:
Data are median (1st quartile - 3rd quartile), mean ± SD. |
|
Acknowledgments:
This study was possible thanks to the collaboration with the UNIMOL researchers and healthcare professionals at the Faculty of Medicine of the Universidad de Cartagena.
References
- Berg AH, Combs TP, Scherer PE. ACRP30/adiponectin: an adipokine regulating glucose and lipid metabolism. Trends Endocrinol Metab 2002. 13(2):84-89. [DOD] [CrossRef]
- Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem 1995. 270(45):26746-26749. [DOD] [CrossRef]
- Vasiliauskaite-Brooks I, Sounier R, Rochaix P, Bellot G, Fortier M, Hoh F, De Colibus L, Bechara C, Saied EM, Arenz C, et al. Structural insights into adiponectin receptors suggest ceramidase activity. Nature 2017. In press. [DOD]
- Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 2003. 423(6941):762-769. [DOD] [CrossRef]
- Tanabe H, Fujii Y, Okada-Iwabu M, Iwabu M, Nakamura Y, Hosaka T, Motoyama K, Ikeda M, Wakiyama M, Terada T, et al. Crystal structures of the human adiponectin receptors. Nature 2015. 520(7547):312-316. [DOD] [CrossRef]
- Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S, Yamashita S, Noda M, Kita S, Ueki K, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 2002. 8(11):1288-1295. [DOD] [CrossRef]
- Michal G, Shomburg D. Biochemical pathways: an atlas of biochemistry and molecular biology. Boehringer Mannheim 2012. [DOD]
- Valsangkar DS, Downs SM. Acetyl CoA carboxylase inactivation and meiotic maturation in mouse oocytes. Mol Reprod Dev 2015. 82(9):679-693. [DOD] [CrossRef]
- Combs TP, Marliss EB. Adiponectin signaling in the liver. Rev Endocr Metab Disord 2014. 15(2):137-147. [DOD] [CrossRef]
- Du Y, Li R, Lau WB, Zhao J, Lopez B, Christopher TA, Ma XL, Wang Y. Adiponectin at physiologically relevant concentrations enhances the vasorelaxative effect of acetylcholine via Cav-1/AdipoR-1 signaling. Plos One 2016. 11(3):e0152247. [DOD] [CrossRef]
- Sun J, Gao Y, Yao T, Huang Y, He Z, Kong X, Yu KJ, Wang RT, Guo H, Yan J, et al. Adiponectin potentiates the acute effects of leptin in arcuate Pomc neurons. Mol Metab 2016. 5(10):882-891. [DOD] [CrossRef]
- Tilija Pun N, Park PH. Role of p62 in the suppression of inflammatory cytokine production by adiponectin in macrophages: Involvement of autophagy and p21/Nrf2 axis. Sci Rep 2017. 7(1):393. [DOD] [CrossRef]
- Ruchat SM, Loos RJ, Rankinen T, Vohl MC, Weisnagel SJ, Despres JP, Bouchard C, Perusse L. Associations between glucose tolerance, insulin sensitivity and insulin secretion phenotypes and polymorphisms in adiponectin and adiponectin receptor genes in the Quebec Family Study. Diabet Med 2008. 25(4):400-406. [DOD] [CrossRef]
- Loos RJ, Ruchat S, Rankinen T, Tremblay A, Perusse L, Bouchard C. Adiponectin and adiponectin receptor gene variants in relation to resting metabolic rate, respiratory quotient, and adiposity-related phenotypes in the Quebec Family Study. Am J Clin Nutr 2007. 85(1):26-34. [DOD]
- Namvaran F, Rahimi-Moghaddam P, Azarpira N, Dabbaghmanesh MH. Polymorphism of adiponectin (45T/G) and adiponectin receptor-2 (795G/A) in an Iranian population: relation with insulin resistance and response to treatment with pioglitazone in patients with type 2 diabetes mellitus. Mol Biol Rep 2012. 39(5):5511-5518. [DOD] [CrossRef]
- Broedl UC, Lehrke M, Fleischer-Brielmaier E, Tietz AB, Nagel JM, Goke B, Lohse P, Parhofer KG. Genetic variants of adiponectin receptor 2 are associated with increased adiponectin levels and decreased triglyceride/VLDL levels in patients with metabolic syndrome. Cardiovasc Diabetol 2006. 5:11. [DOD] [CrossRef]
- Beckers S, de Freitas F, Zegers D, Mertens IL, Verrijken A, Van Camp JK, Van Gaal LF, Van Hul W. No conclusive evidence for association of polymorphisms in the adiponectin receptor 1 gene, AdipoR1, with common obesity. Endocrine 2013. 43(1):120-126. [DOD] [CrossRef]
- Peters KE, Beilby J, Cadby G, Warrington NM, Bruce DG, Davis WA, Davis TM, Wiltshire S, Knuiman M, McQuillan BM, et al. A comprehensive investigation of variants in genes encoding adiponectin (ADIPOQ) and its receptors (ADIPOR1/R2), and their association with serum adiponectin, type 2 diabetes, insulin resistance and the metabolic syndrome. BMC Med Genet 2013. 14:15. [DOD] [CrossRef]
- Schargrodsky H, Hernandez-Hernandez R, Champagne BM, Silva H, Vinueza R, Silva Aycaguer LC, Touboul PJ, Boissonnet CP, Escobedo J, Pellegrini F, et al. CARMELA: assessment of cardiovascular risk in seven Latin American cities. Am J Med 2008. 121(1):58-65. [DOD] [CrossRef]
- Miranda JJ, Herrera VM, Chirinos JA, Gomez LF, Perel P, Pichardo R, Gonzalez A, Sanchez JR, Ferreccio C, Aguilera X, et al. Major cardiovascular risk factors in Latin America: a comparison with the United States. The Latin American Consortium of Studies in Obesity (LASO). Plos One 2013. 8(1):e54056. [DOD] [CrossRef]
- Thornton TA, Bermejo JL. Local and global ancestry inference and applications to genetic association analysis for admixed populations. Genet Epidemiol 2014. 38(Suppl 1):S5-S12. [DOD] [CrossRef]
- Departamento Administrativo Nacional de Estadistica. Censo General de 2005. Libro Censo General. Libro Censo General, 2006. [DOD]
- Gomez Camargo D, Camacho-Mejorado R, Gomez Alegria C, Alario Bello A, Hernandez-Tobias EA, Mora Garcia G, Meraz-Rios MA, Gomez R. Genetic structure of Cartagena de Indias population using hypervariable markers of Y chromosome. Open J Genet 2015. 5(1):1-20. [DOD] [CrossRef]
- Vergara C, Murray T, Rafaels N, Lewis R, Campbell M, Foster C, Gao L, Faruque M, Oliveira RR, Carvalho E, et al. African ancestry is a risk factor for asthma and high total IgE levels in African admixed populations. Genet Epidemiol 2013. 37(4):393-401. [DOD] [CrossRef]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003. 19(1):149-150. [DOD] [CrossRef]
- Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform 2012. 10(2):117-122. [DOD] [CrossRef]
- World Health Organization. WHO STEPS Surveillance Manual: The WHO STEPwise Approach to Chronic Disease Risk Factor Surveillance. 2005, p. 7. [DOD]
- Alberti KG, Zimmet P, Shaw J. The metabolic syndrome - a new worldwide definition. Lancet 2005. 366(9491):1059-1062. [DOD] [CrossRef]
- James PA, Oparil S, Carter BL, Cushman WC, Dennison-Himmelfarb C, Handler J, Lackland DT, LeFevre ML, MacKenzie TD, Ogedegbe O, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA 2014. 311(5):507-520. [DOD] [CrossRef]
- World Health Organization. Waist circumference and waist–hip ratio: report of a WHO expert consultation, Geneva, 8-11 December 2008. WHO Document Production Services 2011. [DOD]
- World Health Organization. Obesity: Preventing and Managing the Global Epidemic. 1998. [DOD]
- Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009. 120(16):1640-1645. [DOD] [CrossRef]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985. 28(7):412-419. [DOD] [CrossRef]
- American Diabetes Association. Standards of Medical Care in Diabetes-2017: Summary of Revisions. Diabetes Care 2017. 40(Suppl 1):S4-S5. [DOD]
- Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature 2012. 491(7422):56-65. [DOD] [CrossRef]
- Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmuller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 2015. 31(8):1334-1336. [DOD] [CrossRef]
- Siitonen N, Pulkkinen L, Lindstrom J, Kolehmainen M, Schwab U, Eriksson JG, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Tuomilehto J, Uusitupa M. Association of ADIPOR2 gene variants with cardiovascular disease and type 2 diabetes risk in individuals with impaired glucose tolerance: the Finnish Diabetes Prevention Study. Cardiovasc Diabetol 2011. 10:83. [DOD] [CrossRef]
- Basson JJ, de Las Fuentes L, Rao DC. Single nucleotide polymorphism-single nucleotide polymorphism interactions among inflammation genes in the genetic architecture of blood pressure in the Framingham Heart Study. Am J Hypertens 2015. 28(2):248-255. [DOD] [CrossRef]
- Collins SC, Luan J, Thompson AJ, Daly A, Semple RK, O'Rahilly S, Wareham NJ, Barroso I. Adiponectin receptor genes: mutation screening in syndromes of insulin resistance and association studies for type 2 diabetes and metabolic traits in UK populations. Diabetologia 2007. 50(3):555-562. [DOD] [CrossRef]
- Potapov VA, Chistiakov DA, Dubinina A, Shamkhalova MS, Shestakova MV, Nosikov VV. Adiponectin and adiponectin receptor gene variants in relation to type 2 diabetes and insulin resistance-related phenotypes. Rev Diabet Stud 2008. 5(1):28-37. [DOD] [CrossRef]
- Mather KJ, Christophi CA, Jablonski KA, Knowler WC, Goldberg RB, Kahn SE, Spector T, Dastani Z, Waterworth D, Richards JB, et al. Common variants in genes encoding adiponectin (ADIPOQ) and its receptors (ADIPOR1/2), adiponectin concentrations, and diabetes incidence in the Diabetes Prevention Program. Diabet Med 2012. 29(12):1579-1588. [DOD] [CrossRef]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010. 10(3):564-567. [DOD] [CrossRef]
- Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc 2011. 6(2):121-133. [DOD] [CrossRef]
- R Development Core Team. R: A language and environment for statistical computing. 2016, R Foundation for Statistical Computing: Viena, Austria. [DOD]
- Kundu S, Aulchenko YS, van Duijn CM, Janssens AC. PredictABEL: an R package for the assessment of risk prediction models. Eur J Epidemiol 2011. 26(4):261-264. [DOD] [CrossRef]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet 2000. 67(1):170-181. [DOD] [CrossRef]
- Builes JJ, Martinez B, Gomez A, Caraballo L, Espinal C, Aguirre D, Montoya A, Moreno M, Amorim A, Gusmao L, Bravo ML. Y chromosome STR haplotypes in the Caribbean city of Cartagena (Colombia). Forensic Sci Int 2007. 167(1):62-69. [DOD] [CrossRef]
- Noguera MC, Schwegler A, Gomes V, Briceno I, Alvarez L, Uricoechea D, Amorim A, Benavides E, Silvera C, Charris M, et al. Colombia's racial crucible: Y chromosome evidence from six admixed communities in the Department of Bolivar. Ann Hum Biol 2014. 41(5):453-459. [DOD] [CrossRef]
- Departamento Administrativo Distrital de Salud. Perfil Epidemiologico de Cartagena de Indias. Alcaldia de Cartagena de Indias 2012. [DOD]
- Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, et al. STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. Plos Med 2009. 6(2):e22. [DOD] [CrossRef]
- Bermudez VJ, Rojas E, Toledo A, Rodriguez-Molina D, Vega K, Suarez L, Pacheco M, Canelon R, Arraiz N, Rojas J, Velasco M. Single-nucleotide polymorphisms in adiponectin, AdipoR1, and AdipoR2 genes: insulin resistance and type 2 diabetes mellitus candidate genes. Am J Ther 2013. 20(4):414-421. [DOD] [CrossRef]
- Kim JT, Kim Y, Cho YM, Koo BK, Lee EK, Shin HD, Jang HC, Choi JW, Oh B, Park KS. Polymorphisms of ADIPOR1 and ADIPOR2 are associated with phenotypes of type 2 diabetes in Koreans. Clin Endocrinol (Oxf) 2009. 70(1):66-74. [DOD] [CrossRef]
- Jin Z, Pu L, Sun L, Chen W, Nan N, Li H, Zhu H, Yang X, Wang N, Hui J, et al. Identification of susceptibility variants in ADIPOR1 gene associated with type 2 diabetes, coronary artery disease and the comorbidity of type 2 diabetes and coronary artery disease. Plos One 2014. 9(6):e100339. [DOD] [CrossRef]
- Wang F, Suo S, Sun L, Yang J, Yang F, Zhao C, Li X, Yuan L, Yu S, Qi T, et al. Analysis of the relationship between ADIPOR1 variants and the susceptibility of chronic metabolic diseases in a Northeast Han Chinese population. Genet Test Mol Biomarkers 2016. 20(2):81-85. [DOD] [CrossRef]
- Liu Q, Yuan B, Lo KA, Patterson HC, Sun Y, Lodish HF. Adiponectin regulates expression of hepatic genes critical for glucose and lipid metabolism. Proc Natl Acad Sci U S A 2012. 109(36):14568-14573. [DOD] [CrossRef]
- Yamauchi T, Kadowaki T. Adiponectin receptor as a key player in healthy longevity and obesity-related diseases. Cell Metab 2013. 17(2):185-196. [DOD] [CrossRef]
- Okada-Iwabu M, Yamauchi T, Iwabu M, Honma T, Hamagami K, Matsuda K, Yamaguchi M, Tanabe H, Kimura-Someya T, Shirouzu M, et al. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature 2013. 503(7477):493-499. [DOD] [CrossRef]
- Hong K, Lee S, Li R, Yang Y, Tanner MA, Wu J, Hill MA. Adiponectin receptor agonist, AdipoRon, causes vasorelaxation predominantly via a direct smooth muscle action. Microcirculation 2016. 23(3):207-220. [DOD] [CrossRef]
- Muhammad SA, Raza W, Nguyen T, Bai B, Wu X, Chen J. Cellular signaling pathways in insulin resistance systems biology analyses of microarray dataset reveals new drug target gene signatures of type 2 diabetes mellitus. Front Physiol 2017. 8:13. [DOD] [CrossRef]
- Rasmussen-Torvik LJ, Pankow JS, Jacobs DR Jr, Steinberger J, Moran A, Sinaiko AR. The association of SNPs in ADIPOQ, ADIPOR1, and ADIPOR2 with insulin sensitivity in a cohort of adolescents and their parents. Hum Genet 2009. 125(1):21-28. [DOD] [CrossRef]
- Marek G, Pannu V, Shanmugham P, Pancione B, Mascia D, Crosson S, Ishimoto T, Sautin YY. Adiponectin resistance and proinflammatory changes in the visceral adipose tissue induced by fructose consumption via ketohexokinase-dependent pathway. Diabetes 2015. 64(2):508-518. [DOD] [CrossRef]
- Tsuchida A, Yamauchi T, Ito Y, Hada Y, Maki T, Takekawa S, Kamon J, Kobayashi M, Suzuki R, Hara K, et al. Insulin/Foxo1 pathway regulates expression levels of adiponectin receptors and adiponectin sensitivity. J Biol Chem 2004. 279(29):30817-30822. [DOD] [CrossRef]
- Kotronen A, Yki-Jarvinen H, Aminoff A, Bergholm R, Pietilainen KH, Westerbacka J, Talmud PJ, Humphries SE, Hamsten A, Isomaa B, et al. Genetic variation in the ADIPOR2 gene is associated with liver fat content and its surrogate markers in three independent cohorts. Eur J Endocrinol 2009. 160(4):593-602. [DOD] [CrossRef]
- Yeh E, Kimura L, Errera FI, Angeli CB, Mingroni-Netto RC, Silva ME, Canani LH, Passos-Bueno MR. Association of polymorphisms at the ADIPOR1 regulatory region with type 2 diabetes and body mass index in a Brazilian population with European or African ancestry. Braz J Med Biol Res 2008. 41(6):468-472. [DOD] [CrossRef]
- Richardson DK, Schneider J, Fourcaudot MJ, Rodriguez LM, Arya R, Dyer TD, Almasy L, Blangero J, Stern MP, Defronzo RA, et al. Association between variants in the genes for adiponectin and its receptors with insulin resistance syndrome (IRS)-related phenotypes in Mexican Americans. Diabetologia 2006. 49(10):2317-2328. [DOD] [CrossRef]
- Castilhos JK, Sprinz E, Lazzaretti RK, Kuhmmer R, Mattevi VS. Polymorphisms in adiponectin receptor genes are associated with lipodystrophy-related phenotypes in HIV-infected patients receiving antiretroviral therapy. HIV Med 2015. 16(8):494-501. [DOD] [CrossRef]
- Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, Ramos FJ, Fang Q, Christ-Roberts CY, Hong JY, Kim RY, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol 2006. 8(5):516-523. [DOD] [CrossRef]
- Chou IP, Lin YY, Ding ST, Chen CY. Adiponectin receptor 1 enhances fatty acid metabolism and cell survival in palmitate-treated HepG2 cells through the PI3 K/AKT pathway. Eur J Nutr 2014. 53(3):907-917. [DOD] [CrossRef]
- Chen MC, Lee CJ, Yang CF, Chen YC, Wang JH, Hsu BG. Low serum adiponectin level is associated with metabolic syndrome and is an independent marker of peripheral arterial stiffness in hypertensive patients. Diabetol Metab Syndr 2017. 9:49. [DOD] [CrossRef]
- Medina-Urrutia A, Posadas-Romero C, Posadas-Sanchez R, Jorge-Galarza E, Villarreal-Molina T, Gonzalez-Salazar Mdel C, Cardoso-Saldana G, Vargas-Alarcon G, Torres-Tamayo M, Juarez-Rojas JG. Role of adiponectin and free fatty acids on the association between abdominal visceral fat and insulin resistance. Cardiovasc Diabetol 2015. 14:20. [DOD] [CrossRef]
- Matsushita Y, Nakagawa T, Yamamoto S, Kato T, Ouchi T, Kikuchi N, Takahashi Y, Yokoyama T, Mizoue T, Noda M. Adiponectin and visceral fat associate with cardiovascular risk factors. Obesity (Silver Spring) 2014. 22(1):287-291. [DOD] [CrossRef]
- Hanley AJ, Bowden D, Wagenknecht LE, Balasubramanyam A, Langfeld C, Saad MF, Rotter JI, Guo X, Chen YD, Bryer-Ash M, et al. Associations of adiponectin with body fat distribution and insulin sensitivity in nondiabetic Hispanics and African-Americans. J Clin Endocrinol Metab 2007. 92(7):2665-2671. [DOD] [CrossRef]
- Chrusciel P, Sahebkar A, Rembek-Wieliczko M, Serban MC, Ursoniu S, Mikhailidis DP, Jones SR, Mosteoru S, Blaha MJ, Martin SS, et al. Impact of statin therapy on plasma adiponectin concentrations: A systematic review and meta-analysis of 43 randomized controlled trial arms. Atherosclerosis 2016. 253:194-208. [DOD] [CrossRef]
|